Interflex with Continuous Outcomes
continuous.RmdLet’s load the package as well as the simulated toy datasets. Note that s6-s9 are for the tutorial for interflex with discrete outcomes.
library(interflex)
data(interflex)
ls()
#> [1] "app_hma2015" "app_vernby2013" "s1" "s2"
#> [5] "s3" "s4" "s5" "s6"
#> [9] "s7" "s8" "s9"s1 is a case of a dichotomous treatment indicator with linear conditional marginal effects (CME); s2 is a case of a continuous treatment indicator with linear CME; s3 is a case of a dichotomous treatment indicator with nonlinear CME; s4 is a case of a dichotomous treatment indicator, nonlinear CME, with additive two-way fixed effects; and s5 is a case of a discrete treatment indicator, nonlinear CME, with additive two-way fixed effects. s1-s5 are generated using the following code:
set.seed(1234)
n<-200
d1<-sample(c(0,1),n,replace=TRUE) # dichotomous treatment
d2<-rnorm(n,3,1) # continuous treatment
x<-rnorm(n,3,1) # moderator
z<-rnorm(n,3,1) # covariate
e<-rnorm(n,0,1) # error term
## linear marginal effect
y1<-5 - 4 * x - 9 * d1 + 3 * x * d1 + 1 * z + 2 * e
y2<-5 - 4 * x - 9 * d2 + 3 * x * d2 + 1 * z + 2 * e
s1<-cbind.data.frame(Y = y1, D = d1, X = x, Z1 = z)
s2<-cbind.data.frame(Y = y2, D = d2, X = x, Z1 = z)
## quadratic marginal effect
x3 <- runif(n, -3,3) # uniformly distributed moderator
y3 <- d1*(x3^2-2.5) + (1-d1)*(-1*x3^2+2.5) + 1 * z + 2 * e
s3 <- cbind.data.frame(D=d1, X=x3, Y=y3, Z1 = z)
## adding two-way fixed effects
n <- 500
d4 <-sample(c(0,1),n,replace=TRUE) # dichotomous treatment
x4 <- runif(n, -3,3) # uniformly distributed moderator
z4 <- rnorm(n, 3,1) # covariate
alpha <- 20 * rep(rnorm(n/10), each = 10)
xi <- rep(rnorm(10), n/10)
y4 <- d4*(x4^2-2.5) + (1-d4)*(-1*x4^2+2.5) + 1 * z4 +
alpha + xi + 2 * rnorm(n,0,1)
s4 <- cbind.data.frame(D=d4, X=x4, Y=y4, Z1 = z4, unit = rep(1:(n/10), each = 10), year = rep(1:10, (n/10)))
## Multiple treatment arms
n <- 600
# treatment 1
d1 <- sample(c('A','B','C'),n,replace=T)
# moderator
x <- runif(n,min=-3, max = 3)
# covriates
z1 <- rnorm(n,0,3)
z2 <- rnorm(n,0,2)
# error
e <- rnorm(n,0,1)
y1 <- rep(NA,n)
y1[which(d1=='A')] <- -x[which(d1=='A')]
y1[which(d1=='B')] <- (1+x)[which(d1=='B')]
y1[which(d1=='C')] <- (4-x*x-x)[which(d1=='C')]
y1 <- y1 + e + z1 + z2
s5 <- cbind.data.frame(D=d1, X=x, Y=y1, Z1 = z1,Z2 = z2)The implied population CME for the DGPs of s1 and s2 are the implied population CME for the DGPs of s3 and s4 are For s5, if we set treatment “A” as our base category, the implied population CME for group “B” and group “C” are, respectively,
The interflex package ships the following functions: interflex, inter.raw, inter.gam, plot. The functionalities of inter.binning and inter.kernel covered by interflex, but they are still supported for backward compatibility.
Raw plots
The first step of the diagnostics is to plot raw data. We supply the
function interflex with the variable names of the
outcome Y, the treatment D, and the moderator
X. You can also supply labels for these variables. If you
supply a variable name to the weights option, the linear
and LOESS fits will be adjusted based on the weights. Note that the
correlations between covariates Z and Y are
NOT partialed out. We use main to add a title to the plot
and cex.main to adjust its size.
interflex(estimator = "raw",Y = "Y", D = "D", X = "X", data = s1,
weights = NULL, Ylabel = "Outcome",
Dlabel = "Treatment", Xlabel="Moderator",
main = "Raw Plot", cex.main = 1.2, ncols=2)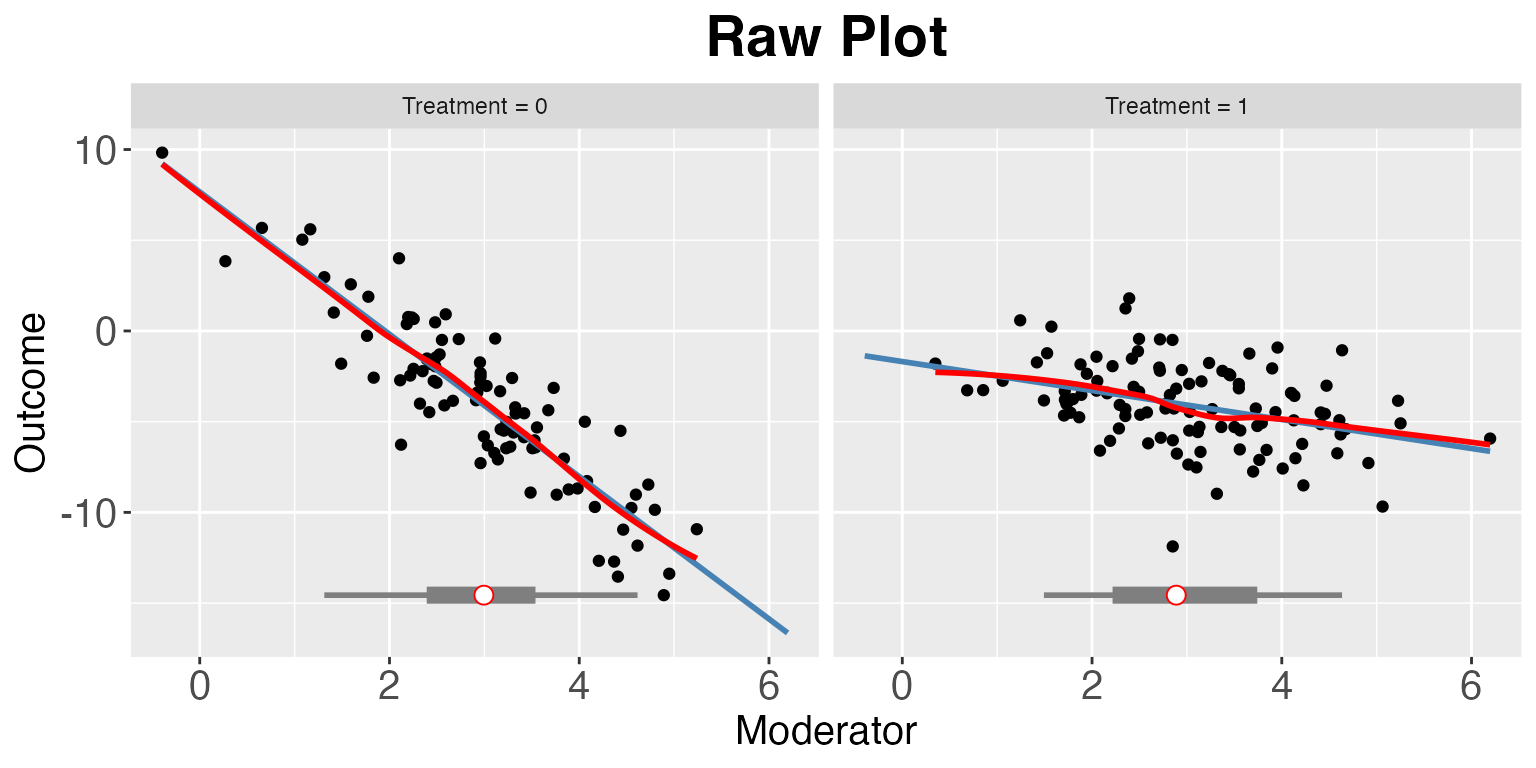
A black-white theme is applied when we set
theme.bw = TRUE. show.grid = FALSE can be used
to remove grid in the plot. Both options are allowed in
interflex and plot.
interflex(estimator = "raw", Y = "Y", D = "D", X = "X", data = s2,
Ylabel = "Outcome", Dlabel = "Treatment", Xlabel="Moderator",
theme.bw = TRUE, show.grid = FALSE, ncols=3)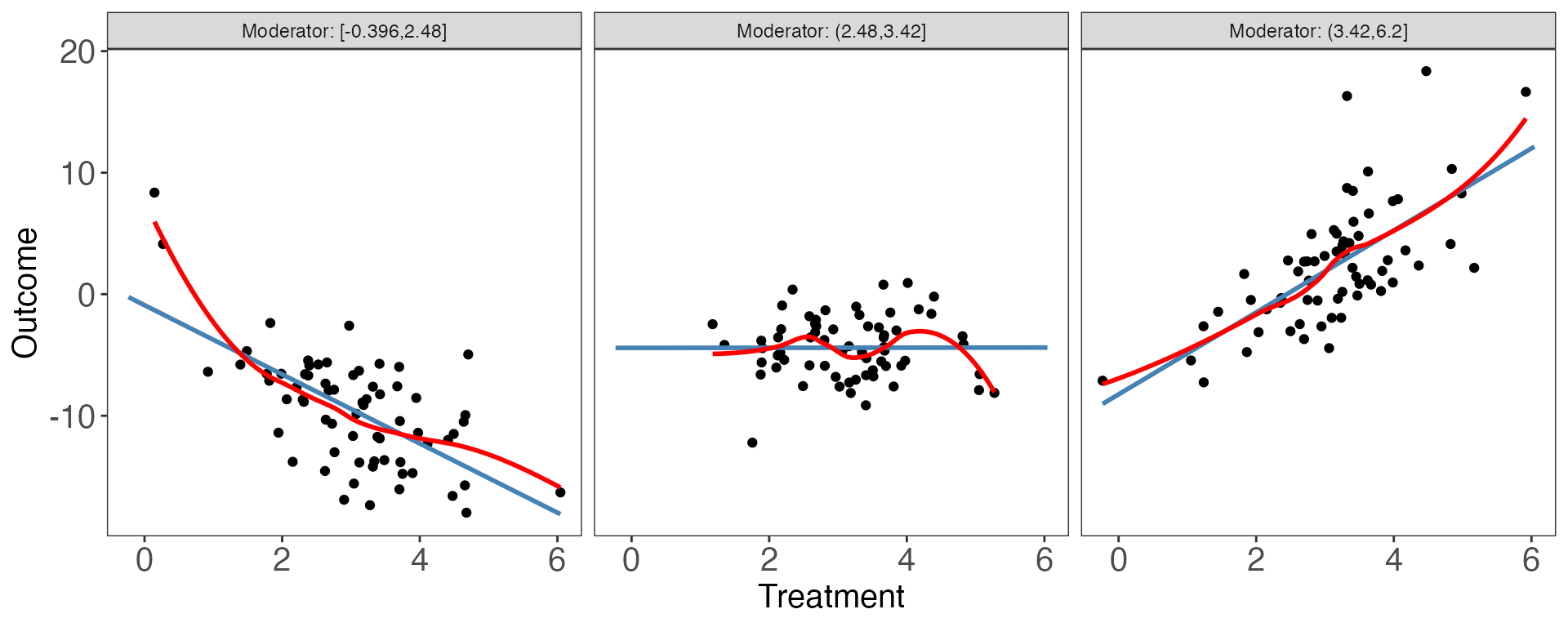
interflex(estimator = "raw", Y = "Y", D = "D", X = "X", data = s3,
Ylabel = "Outcome", Dlabel = "Treatment", Xlabel="Moderator",
ncols=3)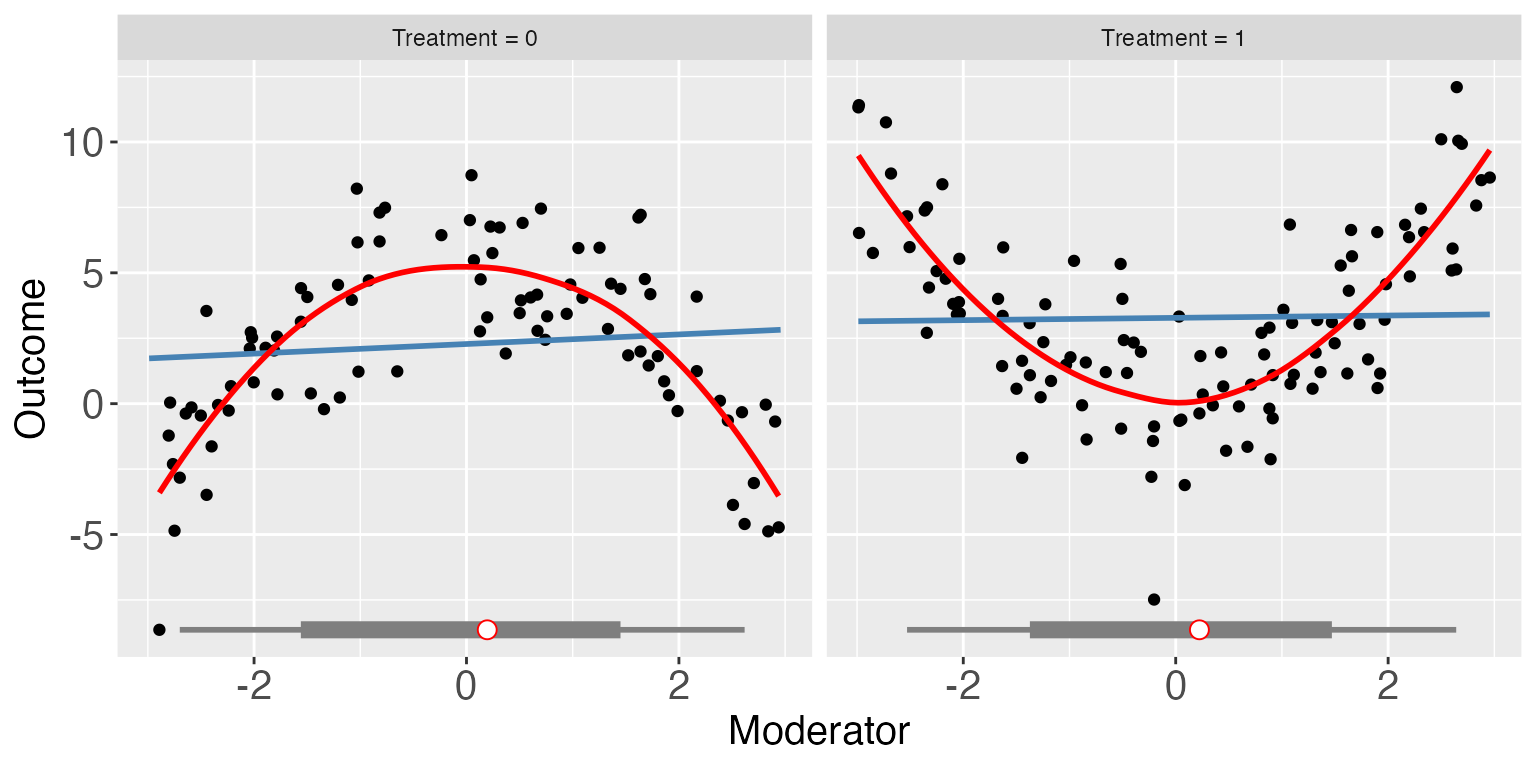
For the continuous treatment case (e.g. s2), we can
also draw a Generalized Additive Model (GAM) plot. You can supply a set
of covariates to be controlled for by supplying Z, which
takes a vector of covariate names (strings).
#>
#> Family: gaussian
#> Link function: identity
#>
#> Formula:
#> Y ~ s(D, X, k = 10) + Z1
#>
#> Estimated degrees of freedom:
#> 8.85 total = 10.85
#>
#> GCV score: 4.59025The binning estimator
The second diagnostic tool is the binning plot. The
nbins option sets the number of bins. The default number of
bins is 3, and equal-sized bins are created based on the distribution of
the moderator. There are four options for the choice of the vcov
estimator: vcov.type = "homoscedastic",
"robust", "cluster",and "pcse".
The default option is "robust".
Note that interflex will also automatically report a
set of statistics when estimator = "binning", including:
(1) the binning estimates and their standard errors and 95% confidence
intervals, (2) the percentage of observations within each bin, (3) the
L-kurtosis of the moderator, and (4) a Wald test to formally test if we
can reject the linear multiplicative interaction model by comparing it
with a more flexible model of multiple bins. .
out <- interflex(Y = "Y", D = "D", X = "X", Z = "Z1", data = s1,
estimator = "binning", vcov.type = "robust",
main = "Marginal Effects", ylim = c(-15, 15))
#> Baseline group not specified; choose treat = 0 as the baseline group.
plot(out)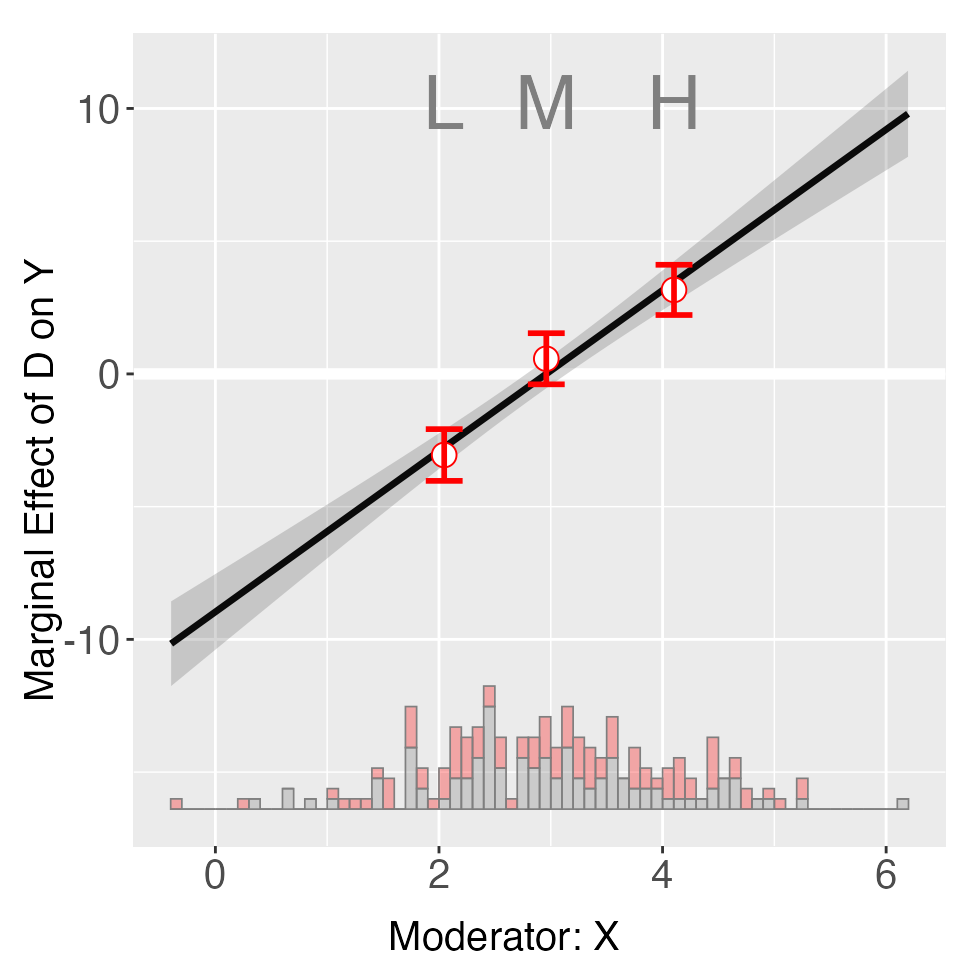
print(out$tests$p.wald)
#> [1] "0.582"We see that the Wald test cannot reject the NULL hypothesis that the
linear interaction model and the three-bin model are statistically
equivalent. If we only want to conduct the linear estimator, we can set
estimator = "linear", We also report the
uniform/simultaneous confidence interval (when setting
vartype = "bootstrap"), as depicted by the dashed
lines.
out2 <- interflex(Y = "Y", D = "D", X = "X", Z = "Z1", data = s1,
estimator = "linear", vcov.type = "robust",
main = "Conditional Marginal Effects", ylim = c(-15, 15))
#> Baseline group not specified; choose treat = 0 as the baseline group.
plot(out2)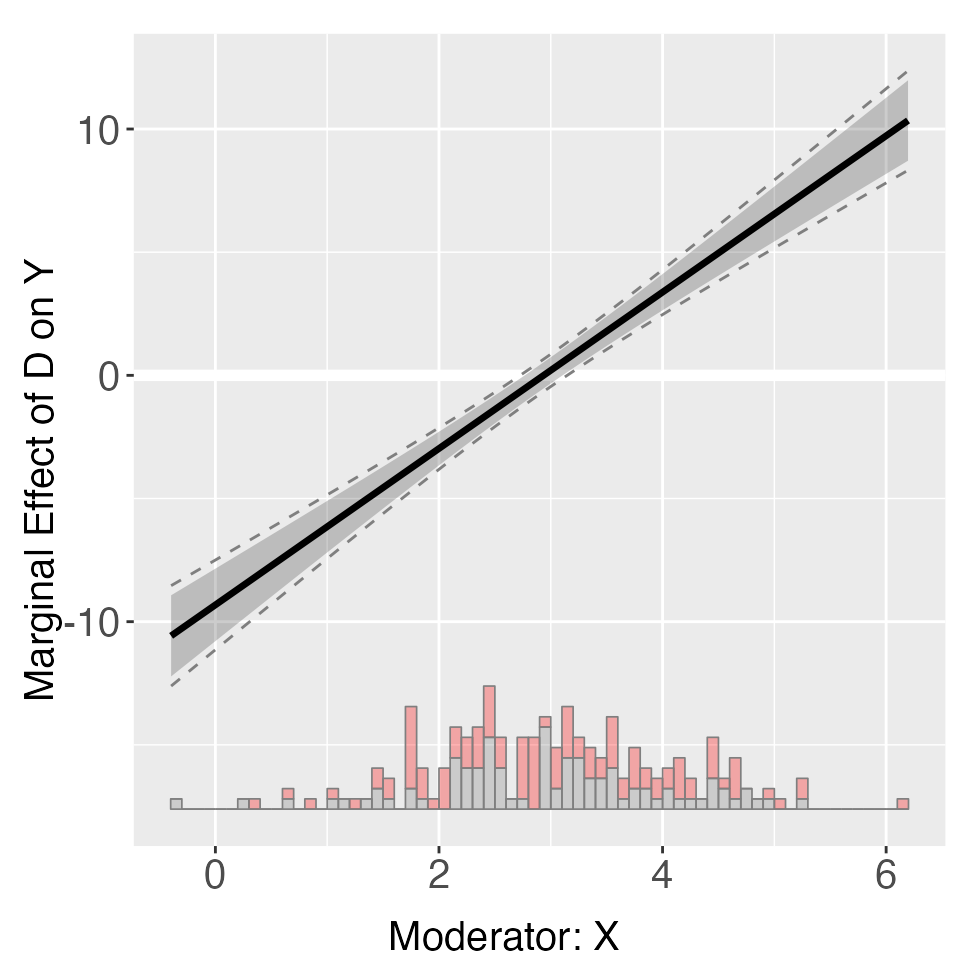
You can access estimates, as well as pointwise and uniform confidence
intervals, from est.lin.
head(out2$est.lin$`1`)
#> X ME sd lower CI(95%) upper CI(95%)
#> [1,] -0.396063535 -10.575601 0.8349589 -12.222311 -8.928892
#> [2,] -0.261533642 -10.148629 0.8046173 -11.735499 -8.561760
#> [3,] -0.127003750 -9.721657 0.7744186 -11.248969 -8.194346
#> [4,] 0.007526143 -9.294685 0.7443803 -10.762755 -7.826616
#> [5,] 0.142056035 -8.867713 0.7145225 -10.276898 -7.458529
#> [6,] 0.276585928 -8.440741 0.6848689 -9.791443 -7.090040
#> lower uniform CI(95%) upper uniform CI(95%)
#> [1,] -12.62133 -8.529875
#> [2,] -12.12002 -8.177242
#> [3,] -11.61905 -7.824260
#> [4,] -11.11849 -7.470885
#> [5,] -10.61836 -7.117067
#> [6,] -10.11873 -6.762749plot allows users to adjust graphic options without
re-estimating the model. The first entry must be a
interflex object. Note that we use
bin.labs = FALSE to hide the label on the top of each bin
and Xdistr = "none" to remove the distribution of the
moderator (not recommended). We use cex.axis and
cex.lab to adjust the font sizes of axis numbers and
labels.
plot(out, xlab = "Moderate is X", Xdistr = "none", bin.labs = FALSE, cex.axis = 0.8, cex.lab = 0.8)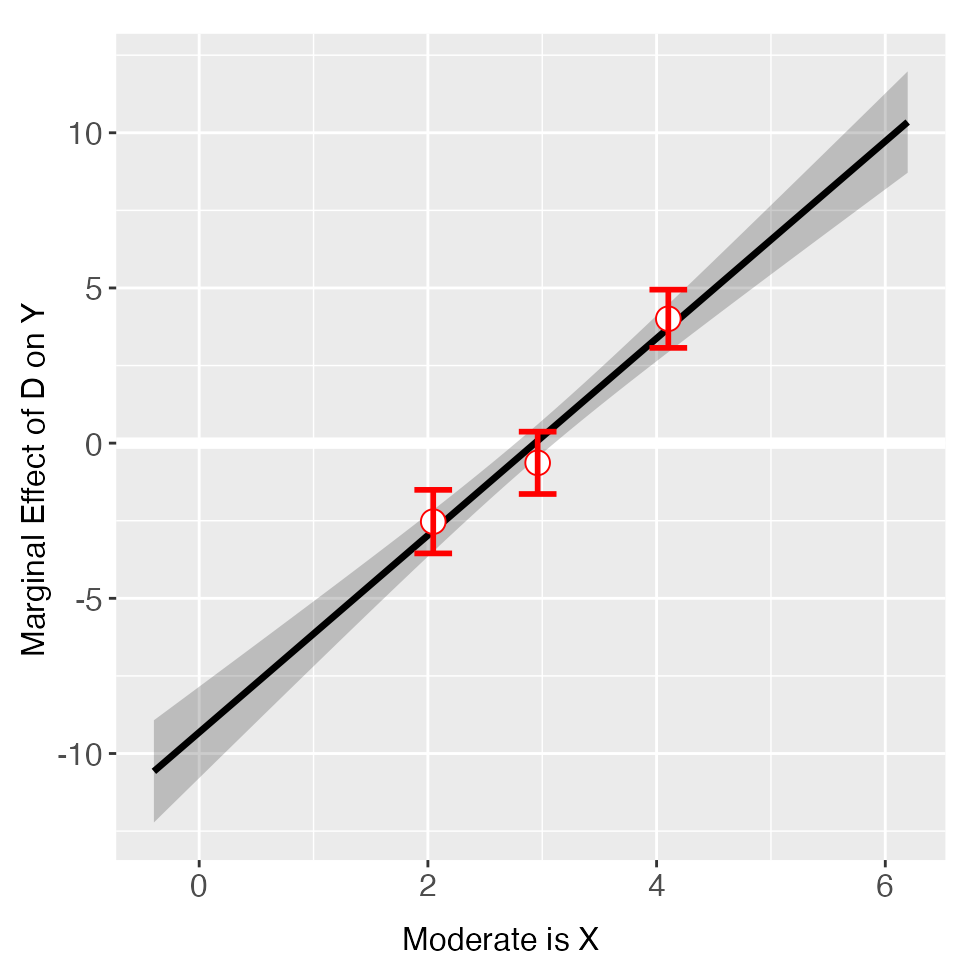
Next, we use Xunif = TRUE to transform the moderator
into a uniformly distributed random variable (based on the rank order in
values of the orginal moderator) before estimating the CME.
nbins = 4 sets the number of bins to 4.
out <- interflex(Y = "Y", D = "D", X = "X", Z = "Z1", data = s1,
estimator = "binning", nbins = 4,
theme.bw = TRUE, Xunif = TRUE)
#> Baseline group not specified; choose treat = 0 as the baseline group.
out$figure
The binning estimates for the continuous case are shown below. We now
present the distribution of the moderator with a density plot using
option Xdist = "density" – the default option is
"hist" or "histogram". We turn off the bin
labels using bin.labs = FALSE.
out <- interflex(Y = "Y", D = "D", X = "X", Z = "Z1", data = s2,
estimator = "binning", Xdistr = "density",
bin.labs = FALSE)
out$figure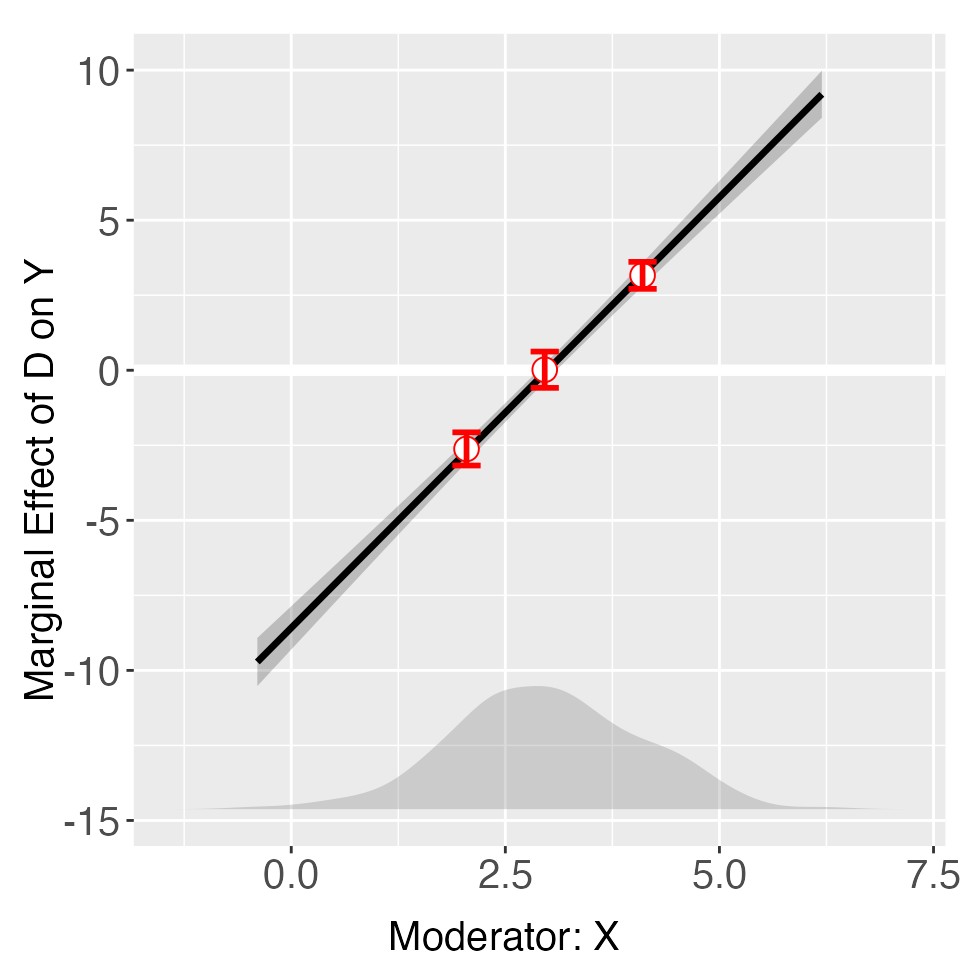
Note that you can customize the cutoff values for the bins, for
example, set cutoffs = c(1, 2, 4, 5) to create five bins:
[minX, 1], (1, 2], (2,4], (4, 5] and (5,maxX] (supplying N numbers will
create N+1 bins). Note that the cutoffs option will
override the nbins option if they are incompatible.
out <- interflex(Y = "Y", D = "D", X = "X", Z = "Z1", data = s2,
estimator = "binning", cutoffs = c(1,2,4,5))
out$figure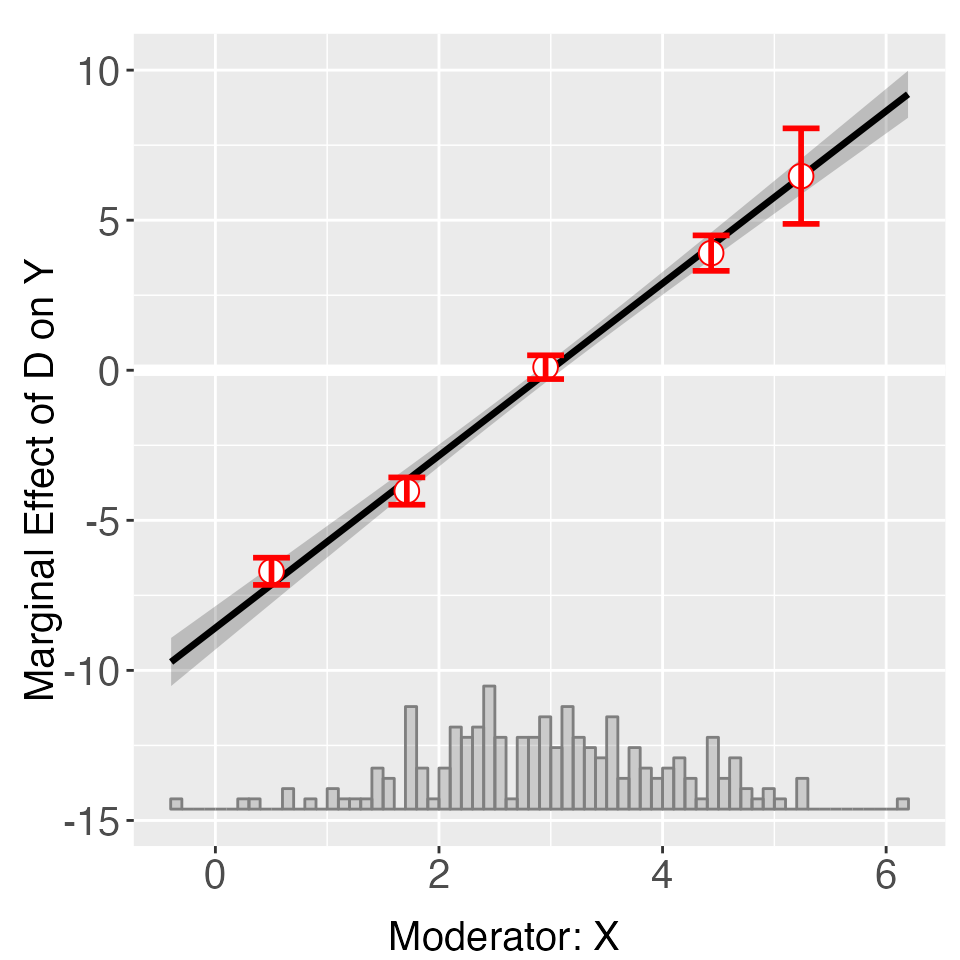
The binning estimates for the dichotomous, nonlinear case
(i.e. s3) are shown below. A linear interaction model
clearly gives misleading CME estimates. The CME plot is stored in
out$figure while the estimates and standard errors are
stored in out$est.linear (linear) and
out$est.bin (binning). The tests results are stored in
out$tests (binning).
out <- interflex(Y = "Y", D = "D", X = "X", Z = "Z1", data = s3,
estimator = "binning")
#> Baseline group not specified; choose treat = 0 as the baseline group.
print(out$tests$p.wald)
#> [1] "0.000"This time the NULL hypothesis that the linear interaction model and the three-bin model are statistically equivalent is safely rejected (p.wald = 0.00).
The kernel estimator
With the kernel method, a bandwidth is first selected via 10-fold least-squares cross-validation.
The standard errors are produced by a non-parametric bootstrap (you
can adjust the number of bootstrap iterations using the
nboots option). The grid option can either be
an integer, which specifies the number of bandwidths to be
cross-validated, or a vector of candidate bandwidths – the default is
grid = 20. You can also specify a clustered group structure
by using the cl option, in which case a block bootstrap
procedure will be performed.
Starting from v.1.1.0, we introduce adaptive bandwidth selection to
interflex. This means that the bandwidth will be
smaller in regions where there are more data and larger in regions where
there are few data points (based on the moderator). When adaptive
bandwidth is being used, bw refers to the bandwidth applied
in the region with the highest density of observations.
Even with an optimized algorithm, the bootstrap procedure for large
datasets can be slow. We incorporate parallel computing
(parallel=TRUE) to speed it up. You can choose the number
of cores to be used for parallel computing; otherwise the
program will detect the number of logical cores in your computer and use
as many cores as possible (warning: this will significantly slow down
your computer!).
out <- interflex(Y = "Y", D = "D", X = "X", Z = "Z1", data = s1, vartype = 'bootstrap',
estimator = "kernel", nboots = 1000,
parallel = TRUE, cores = 31)
#> Baseline group not specified; choose treat = 0 as the baseline group.
#> Cross-validating bandwidth ...
#> Parallel computing with 20 cores...
#> Optimal bw=6.592.
#> Number of evaluation points:50
#> Parallel computing with 20 cores...
#>
out$figure
You can access estimates, as well as pointwise and uniform confidence
intervals, from est.kernel.
head(out$est.kernel$`1`)
#> X TE sd lower CI(95%) upper CI(95%)
#> [1,] -0.396063535 -10.571474 0.8474103 -12.275007 -8.933925
#> [2,] -0.261533642 -10.144602 0.8165569 -11.784214 -8.571760
#> [3,] -0.127003750 -9.717701 0.7858425 -11.293344 -8.209809
#> [4,] 0.007526143 -9.290720 0.7552747 -10.802550 -7.844952
#> [5,] 0.142056035 -8.863691 0.7248648 -10.316085 -7.485393
#> [6,] 0.276585928 -8.436684 0.6946379 -9.836409 -7.127547
#> lower uniform CI(95%) upper uniform CI(95%)
#> [1,] -12.66221 -8.451586
#> [2,] -12.14507 -8.095122
#> [3,] -11.66239 -7.738630
#> [4,] -11.17968 -7.382128
#> [5,] -10.69281 -7.026905
#> [6,] -10.19913 -6.676001If you specify a bandwidth manually (for example, by setting
bw = 1), the cross-validation step will be skipped. The
main option controls the title of the plot while the
xlim and ylim options control the ranges of
the x-axis and y-axis, respectively.
out <- interflex(Y = "Y", D = "D", X = "X", Z = "Z1", data = s1,
estimator = "kernel", nboots = 1000, bw = 1,
main = "The Linear Case", parallel = TRUE, cores = 31,vartype = 'bootstrap',
xlim = c(-0.5,6.5), ylim = c(-15, 15))
out$figure
For dataset s3, the kernel method produces non-linear CME estimates that are much closer to the effects implied by the true DGP.
out <- interflex(Y = "Y", D = "D", X = "X", Z = "Z1", data = s3, vartype = 'bootstrap',
estimator = "kernel", theme.bw = TRUE, nboots = 1000, parallel = TRUE, cores = 31)
out$figure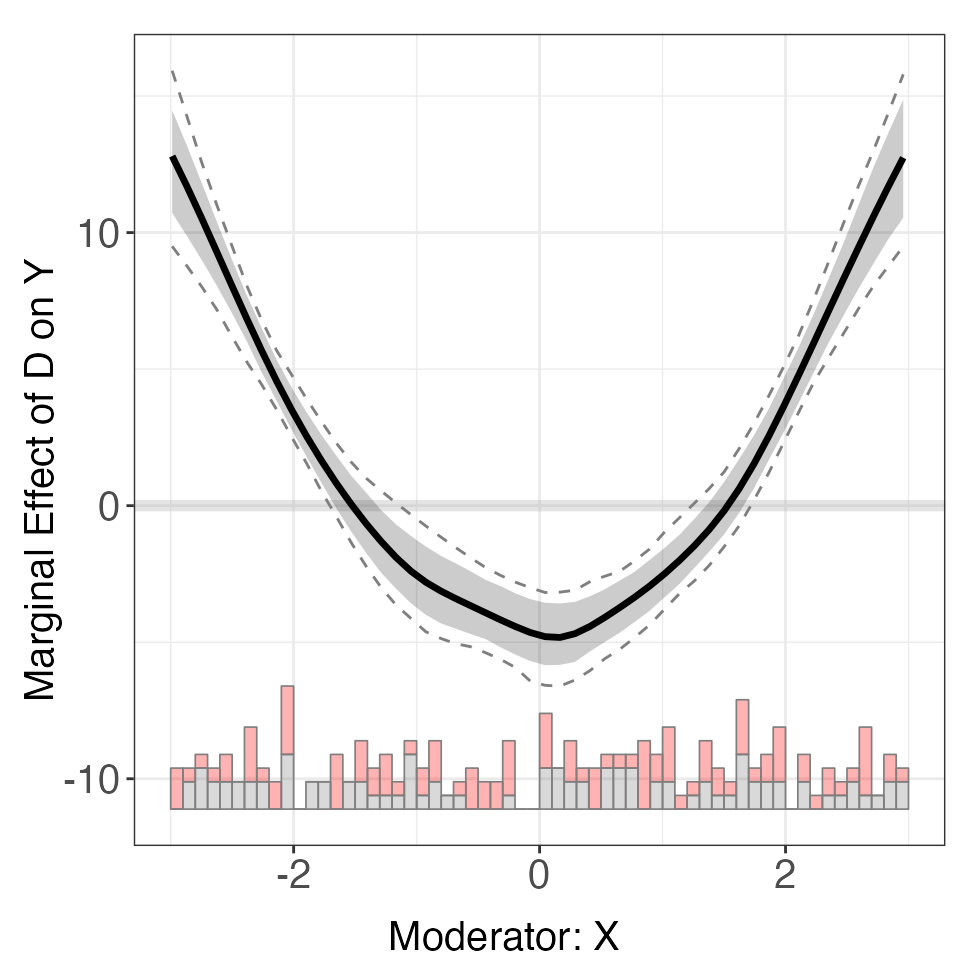
Again, if we only want to change the look of a marginal effect plot,
we can use plot to save time. CI = FALSE
removes the confidence interval ribbon.
plot(out, main = "Nonlinear Conditional Marginal Effects",
ylab = "Coefficients", Xdistr = "density",
xlim = c(-3,3), ylim = c(-10,12), CI = FALSE,
cex.main = 0.8, cex.lab = 0.7, cex.axis = 0.7)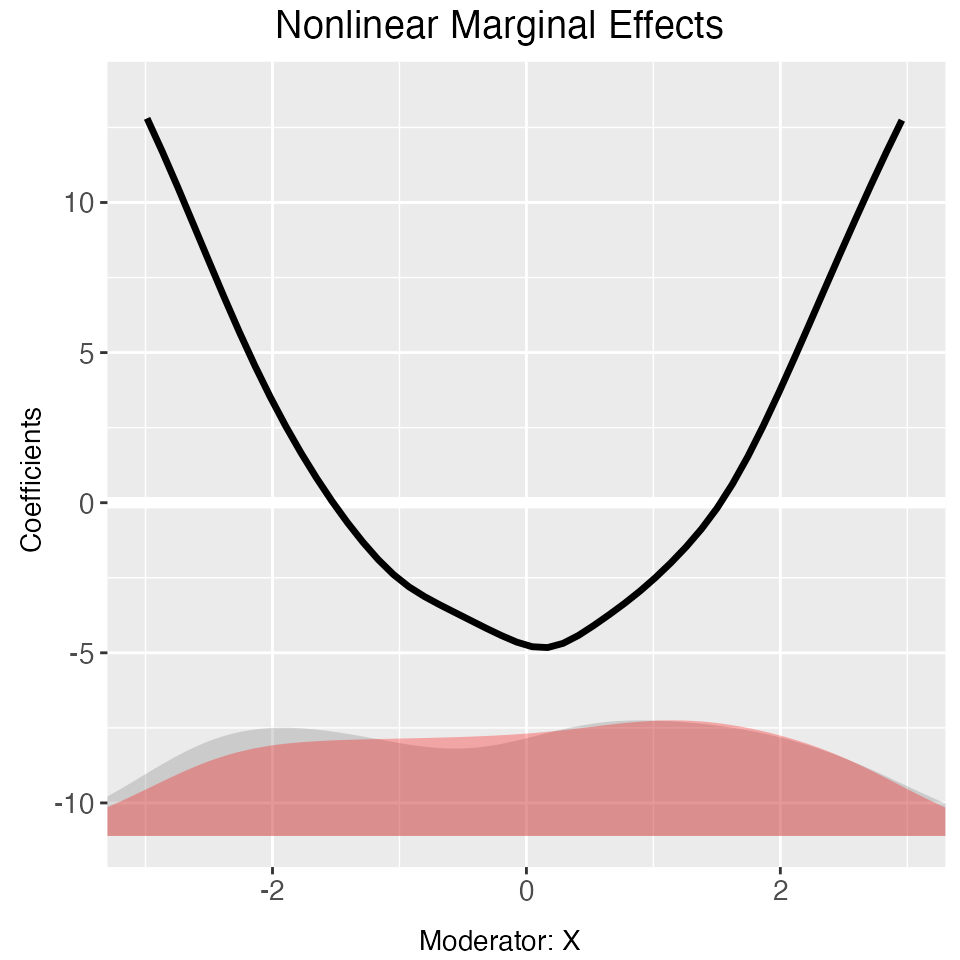
The semi-parametric marginal effect estimates are stored in
out$est.kernel.
Note that we can use the file option to save a plot to a
file in interflex (e.g. by setting
file = "myplot.pdf" or
file = "myplot.png").
Fixed effects
We move on to linear fixed effects models. Remember in s4, a large chunk of the variation in the outcome variable is driven by group fixed effects. Below is a scatterplot of the raw data (group index vs. outcome). Red and green dots represent treatment and control units, respectively. We can see that outcomes are highly correlated within a group.
library(ggplot2)
ggplot() + geom_point(data = s4,aes(x = unit, y = Y, colour = as.factor(D))) +
guides(colour=FALSE) 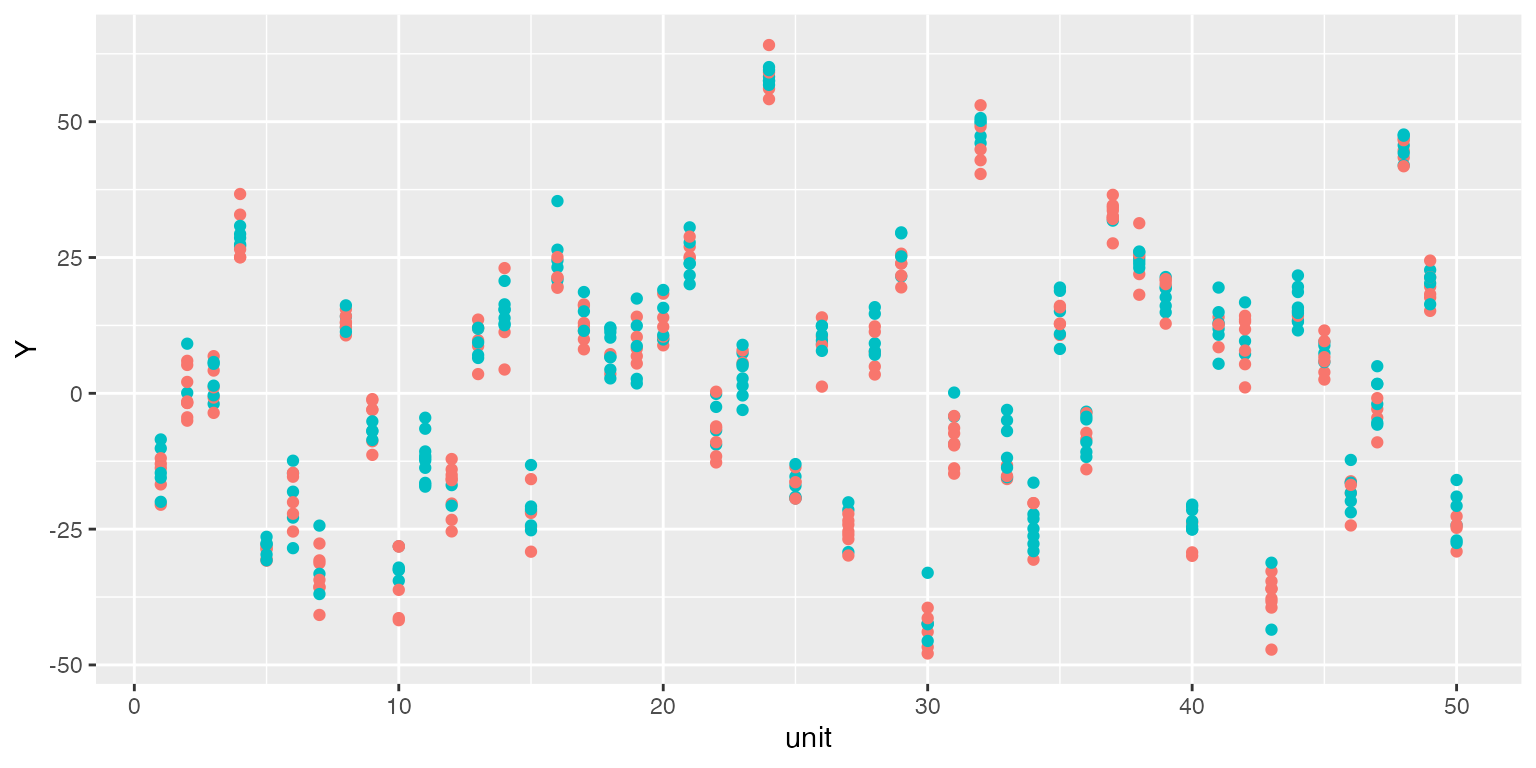
When fixed effects are present, it is possible that we cannot observe a clear pattern of CME in the raw plot as before, while binning estimates have wide confidence intervals:
interflex(estimator = "raw", Y = "Y", D = "D", X = "X", data = s4,
weights = NULL,ncols=2)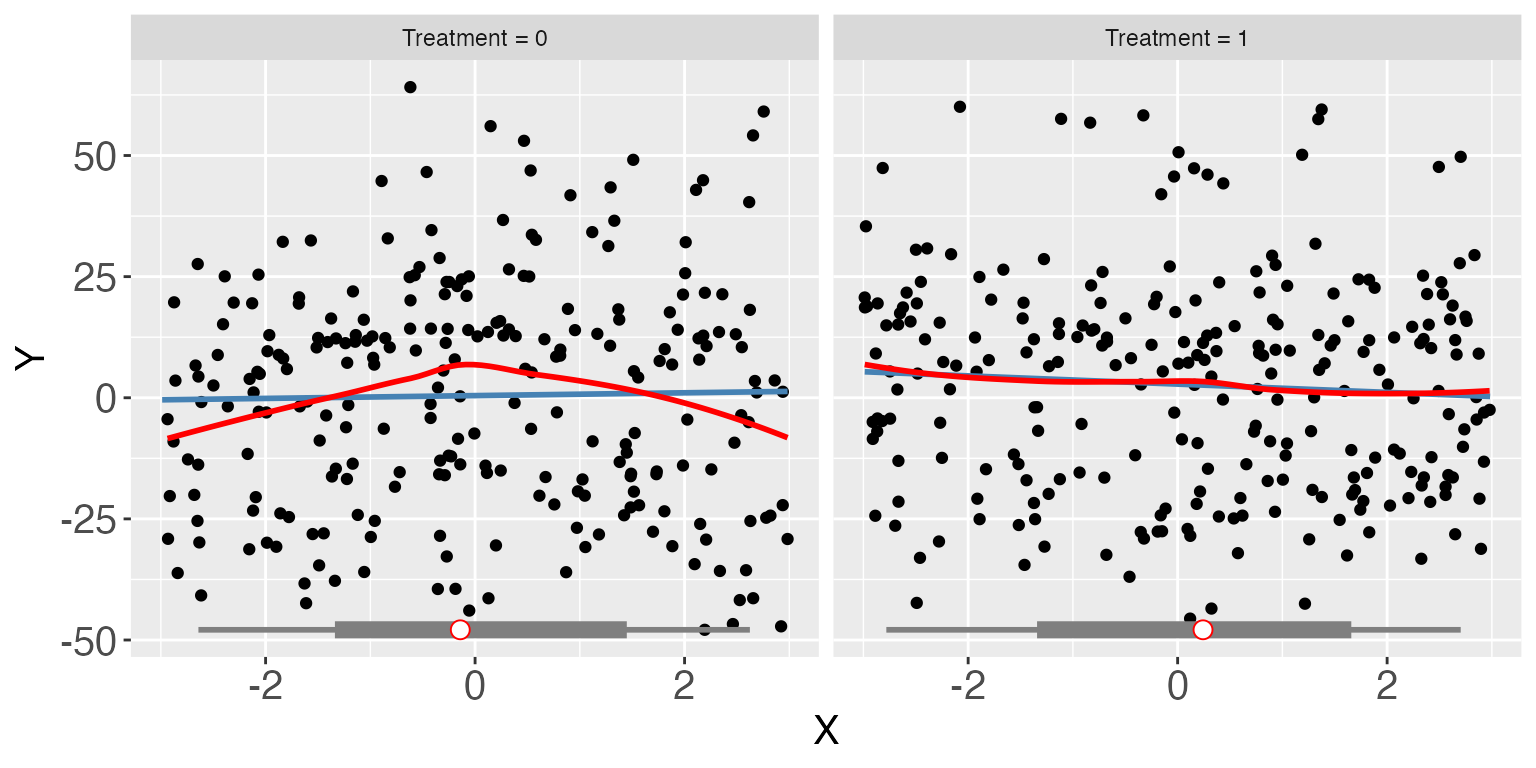
s4.binning <- interflex(Y = "Y", D = "D", X = "X", Z = "Z1", data = s4,
estimator = "binning", FE = NULL, cl = "unit")
plot(s4.binning)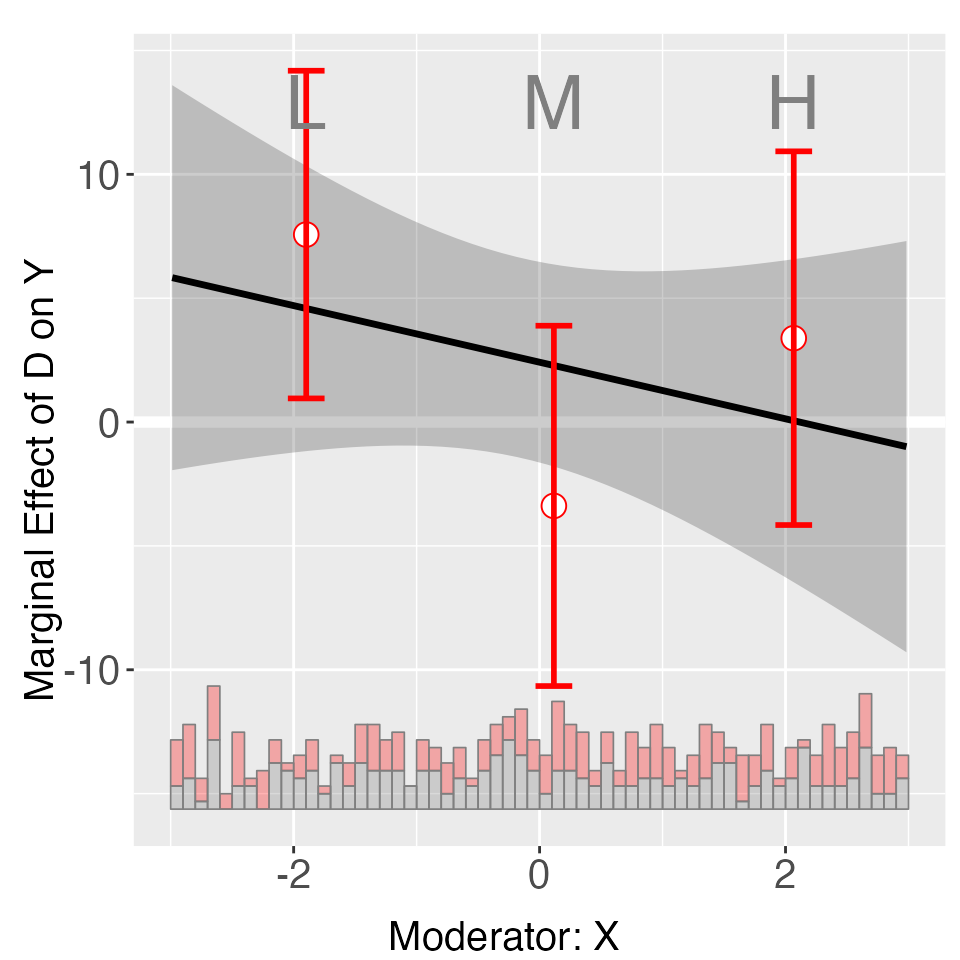
The binning estimates are much more informative when fixed effects
are included, by using the FE option. Note that the number
of group indicators can exceed 2. Our algorithm is optimized for a large
number of fixed effects or many group indicators. The cl
option controls the level at which standard errors are clustered.
s4$wgt <- 1
s4.binning <- interflex(Y = "Y", D = "D", X = "X", Z = "Z1", data = s4,
estimator = "binning", FE = c("unit", "year"),
cl = "unit", weights = "wgt")
plot(s4.binning)
When fixed effects are not taken into account, the kernel estimates are also less precisely estimated. Because the model is incorrectly specified, cross-validated bandwidths also tend to be bigger than optimal.
s4.kernel <- interflex(Y = "Y", D = "D", X = "X", Z = "Z1", data = s4,
estimator = "kernel", FE = NULL, vartype = 'bootstrap',
nboots = 1000, parallel = TRUE, cores = 31,
cl = "unit", weights = "wgt")
plot(s4.kernel)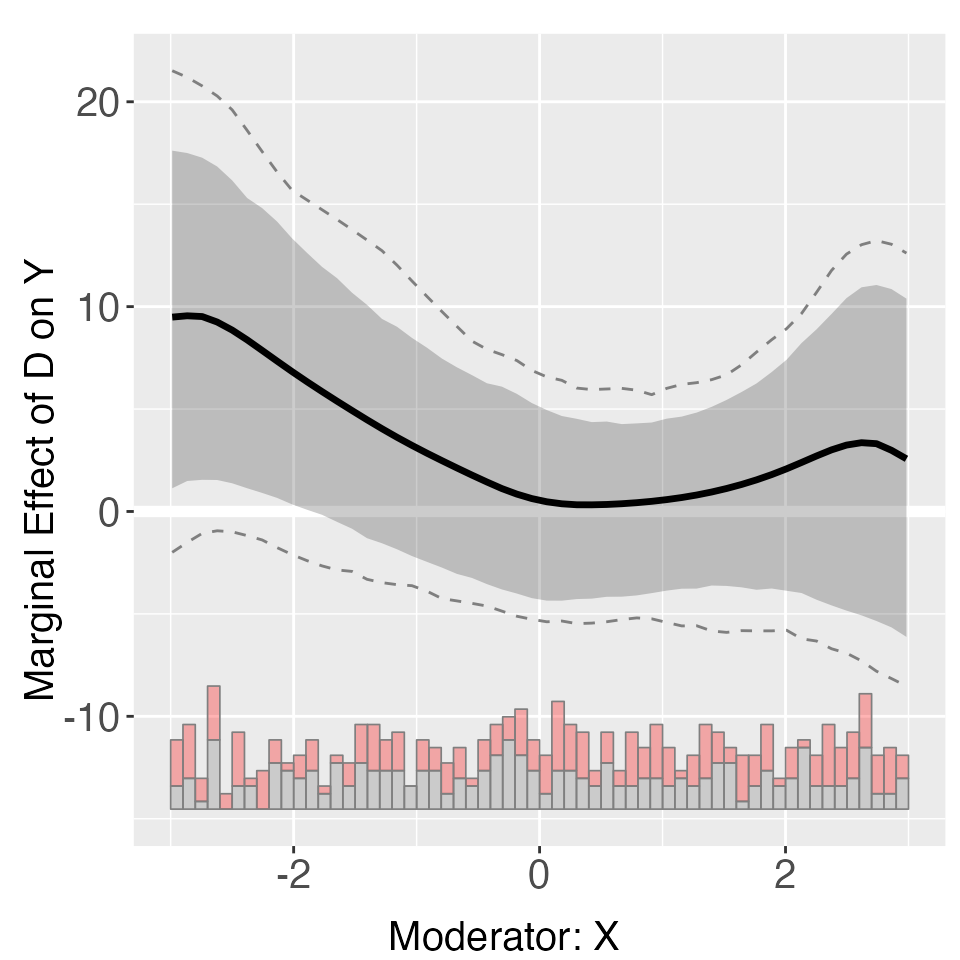
Controlling for fixed effects by using the FE option
solves this problem. The estimates are now much closer to the population
truth. Note that all uncertainty estimates are produced via
bootstrapping. When the cl option is specified, a block
bootstrap procedure will be performed.
s4.kernel <- interflex(Y = "Y", D = "D", X = "X", Z = "Z1", data = s4,
estimator = "kernel", FE = c("unit","year"), cl = "unit",vartype = 'bootstrap',
nboots = 1000, parallel = TRUE, cores = 31,weights = "wgt")
plot(s4.kernel)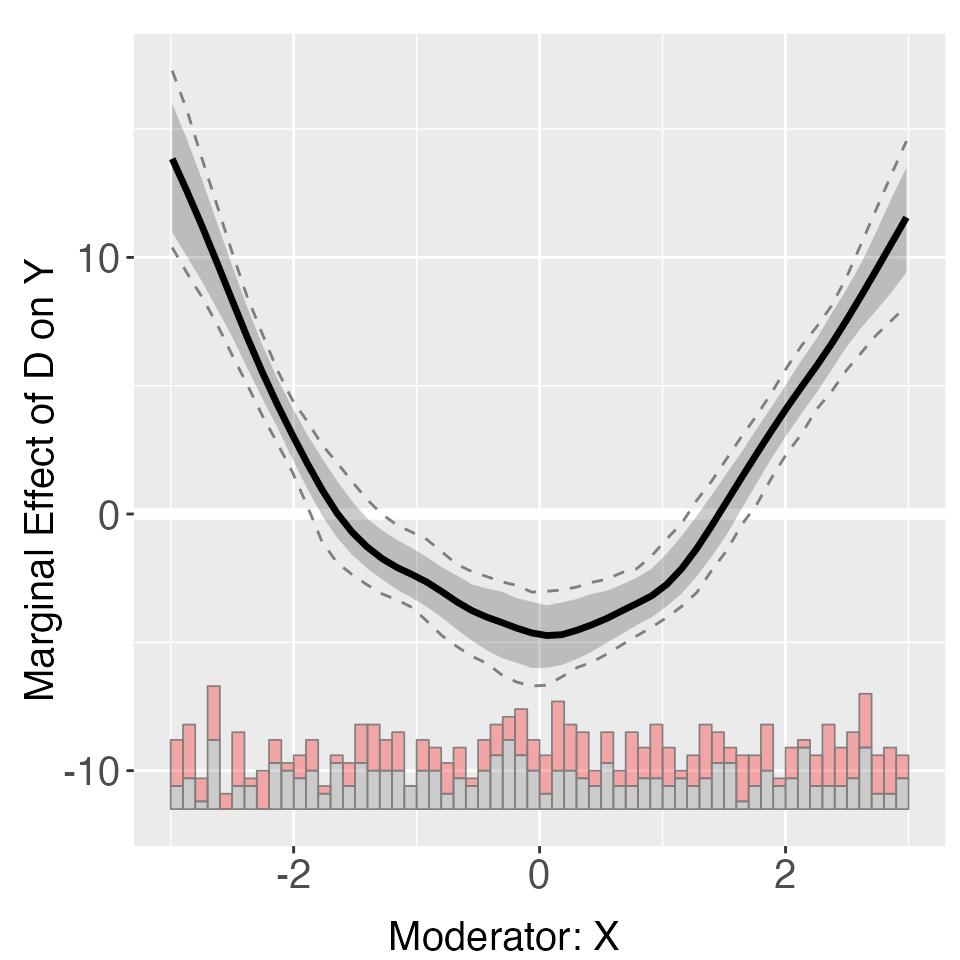
With large datasets, cross-validation or bootstrapping can take a
while. One way to check the result quickly is to shut down the bootstrap
procedure (using CI = FALSE). interflex
will then present the point estimates only. Another way is to supply a
reasonable bandwidth manually by using the bw option such
that cross-validation will be skipped.
s4.kernel <- interflex(Y = "Y", D = "D", X = "X", Z = "Z1", data = s4,
estimator = "kernel", bw = 0.62, FE = c("unit","year"), cl = "unit",
vartype = 'bootstrap',
nboots = 1000, parallel = TRUE, cores = 31,
CI = FALSE, ylim = c(-9, 15), theme.bw = TRUE)
plot(s4.kernel)
Multiple (>2) treatment arms
Next, we will show how interflex can be applied when there are more than two condition. First, we plot the raw data:
interflex(estimator = "raw",Y = "Y", D = "D", X = "X", data = s5, ncols = 3)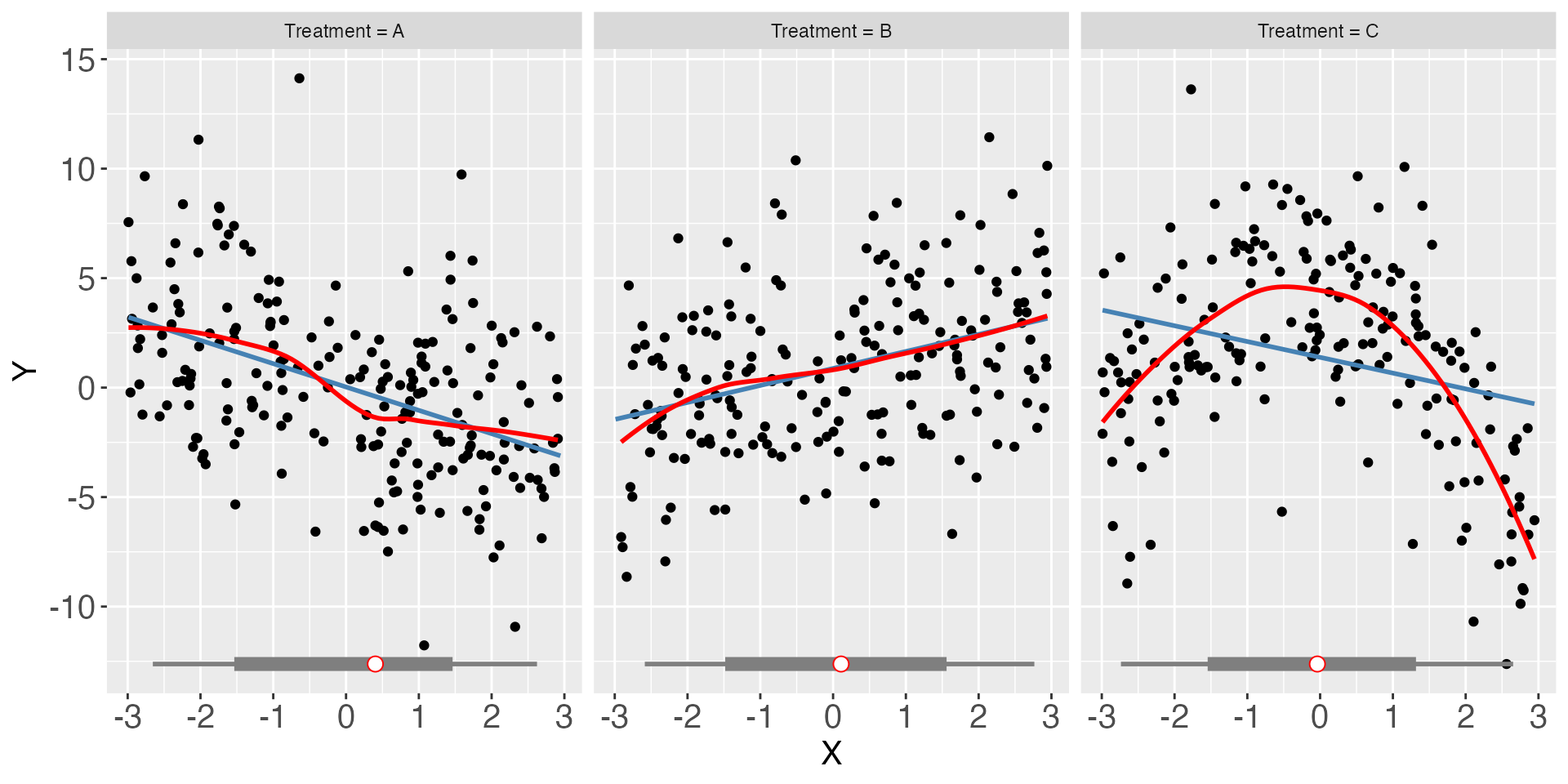
By default, interflex will produce
CME plots, taking one category as the baseline (if not specified, the
base group is selected based on the numeric/character order of the
treatment values). Here we also specify
vartype = "bootstrap" in order to estimate the confidence
intervals.
s5.binning <- interflex(Y = "Y", D = "D", X = "X", Z = c("Z1","Z2"), data = s5,
estimator = "binning", vartype = "bootstrap")
plot(s5.binning)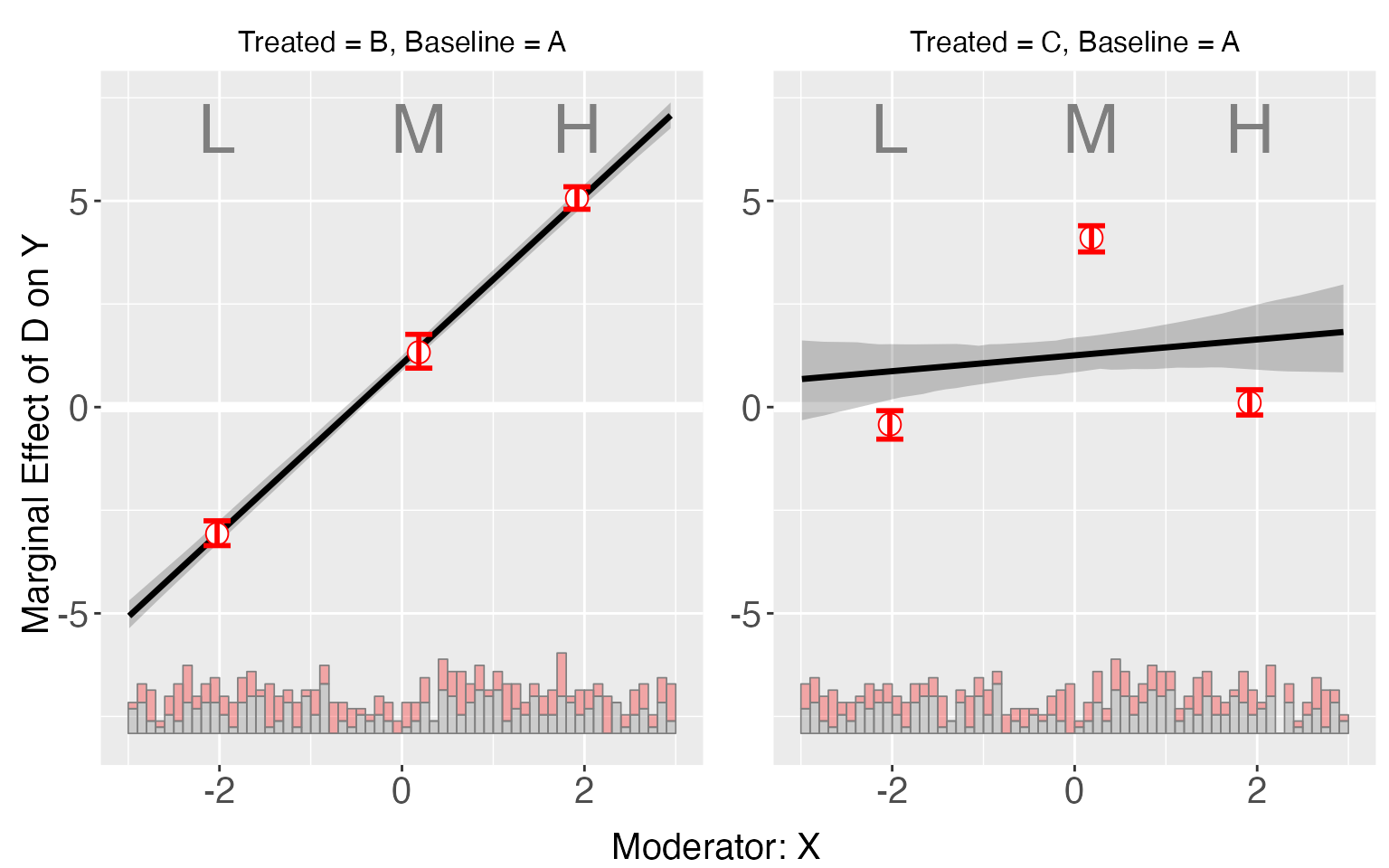
By setting the option order and subtitles
(in either interflex or plot), users
can also specify the order of plots they prefer and the subtitles they
want:
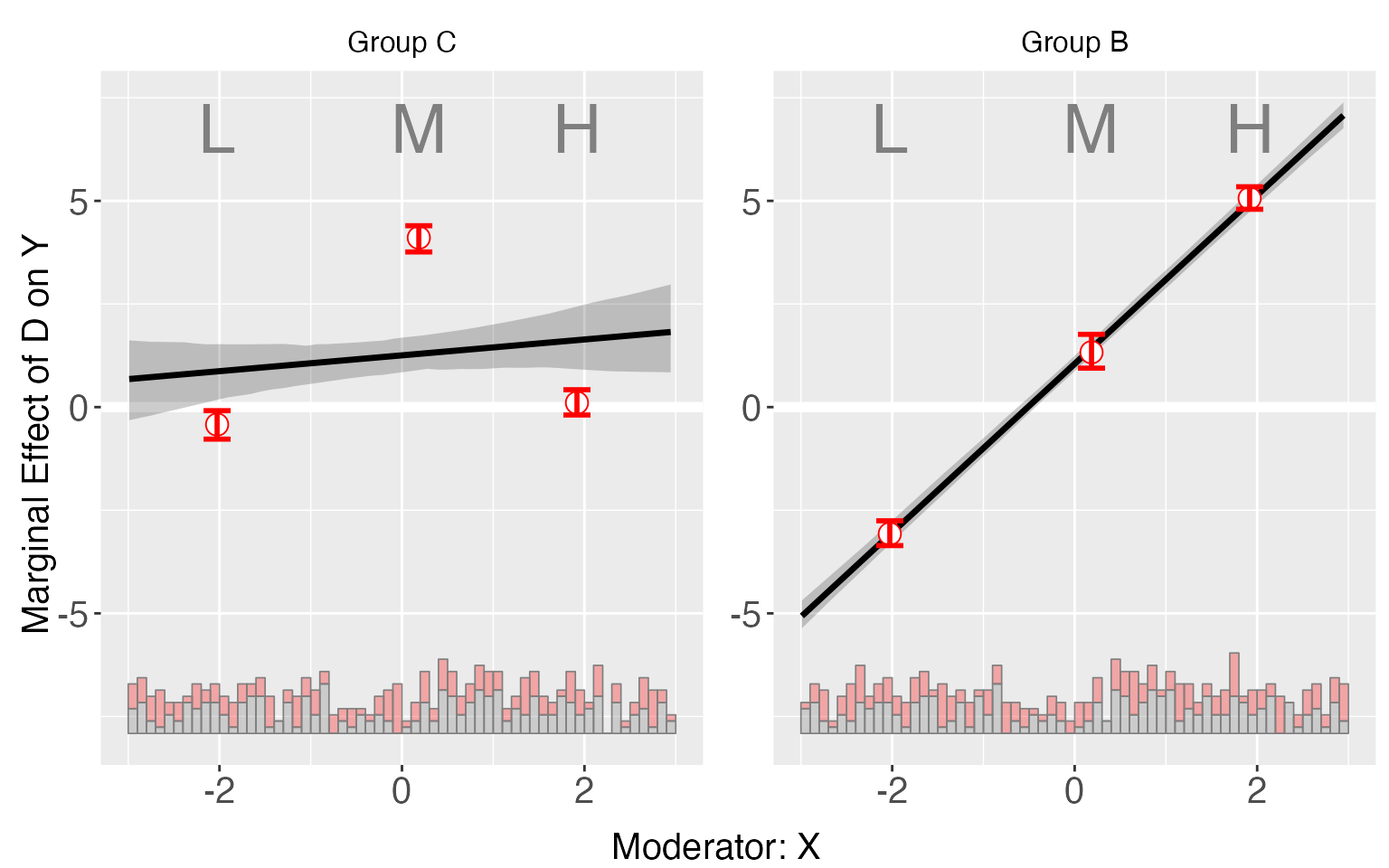
By setting pool = TRUE (in either
interflex or plot), we can combine
these plots. Users can also specify the colors they like, :
plot(s5.binning, order=c("C", "B"), subtitles = c("Control Group","Group C","Group B"),
pool = TRUE, color = c("Salmon","#7FC97F", "#BEAED4"))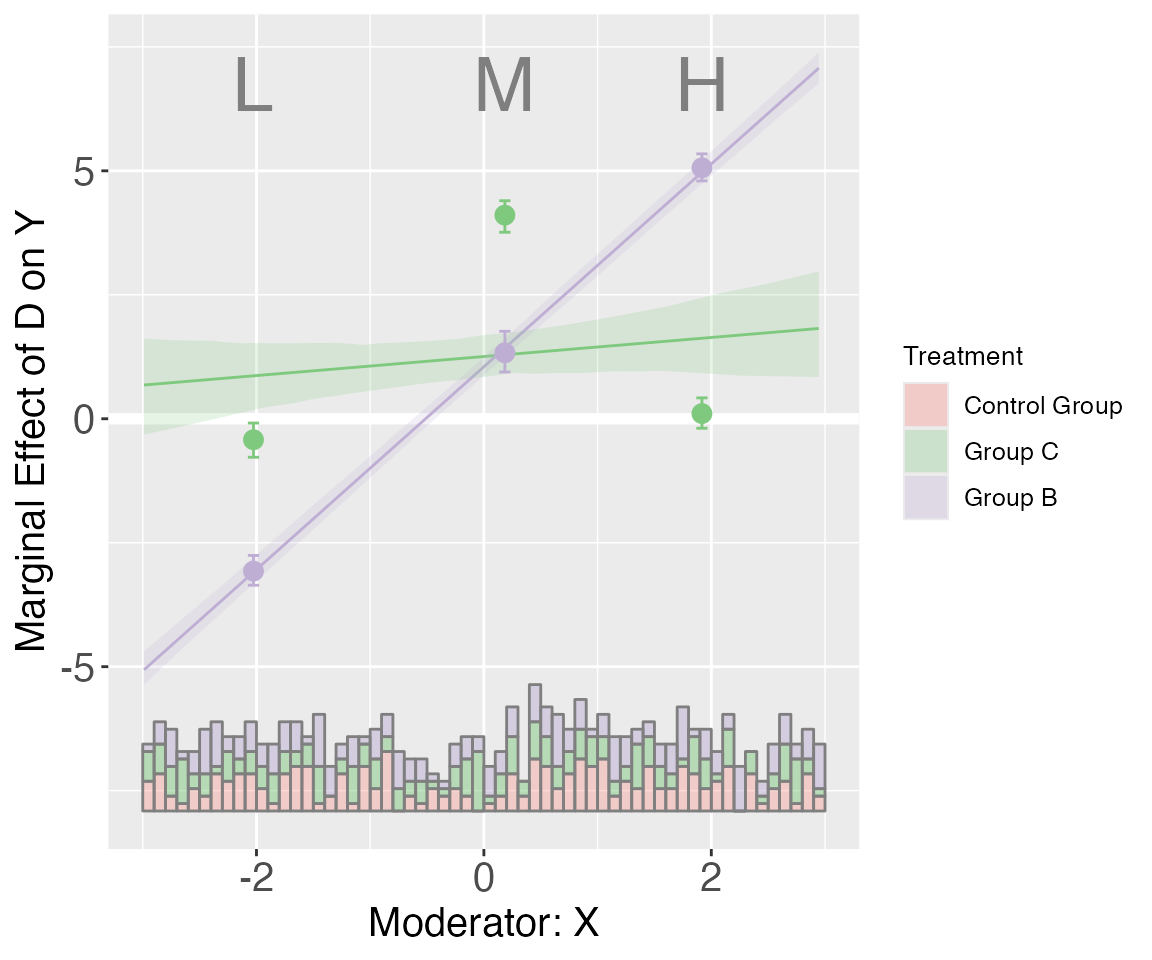
We specify the base group using the option base:
s5.binning2 <- interflex(Y = "Y", D = "D", X = "X",Z = c("Z1", "Z2"), data = s5,
estimator = "binning", base = "B", vartype = "bootstrap")
plot(s5.binning2)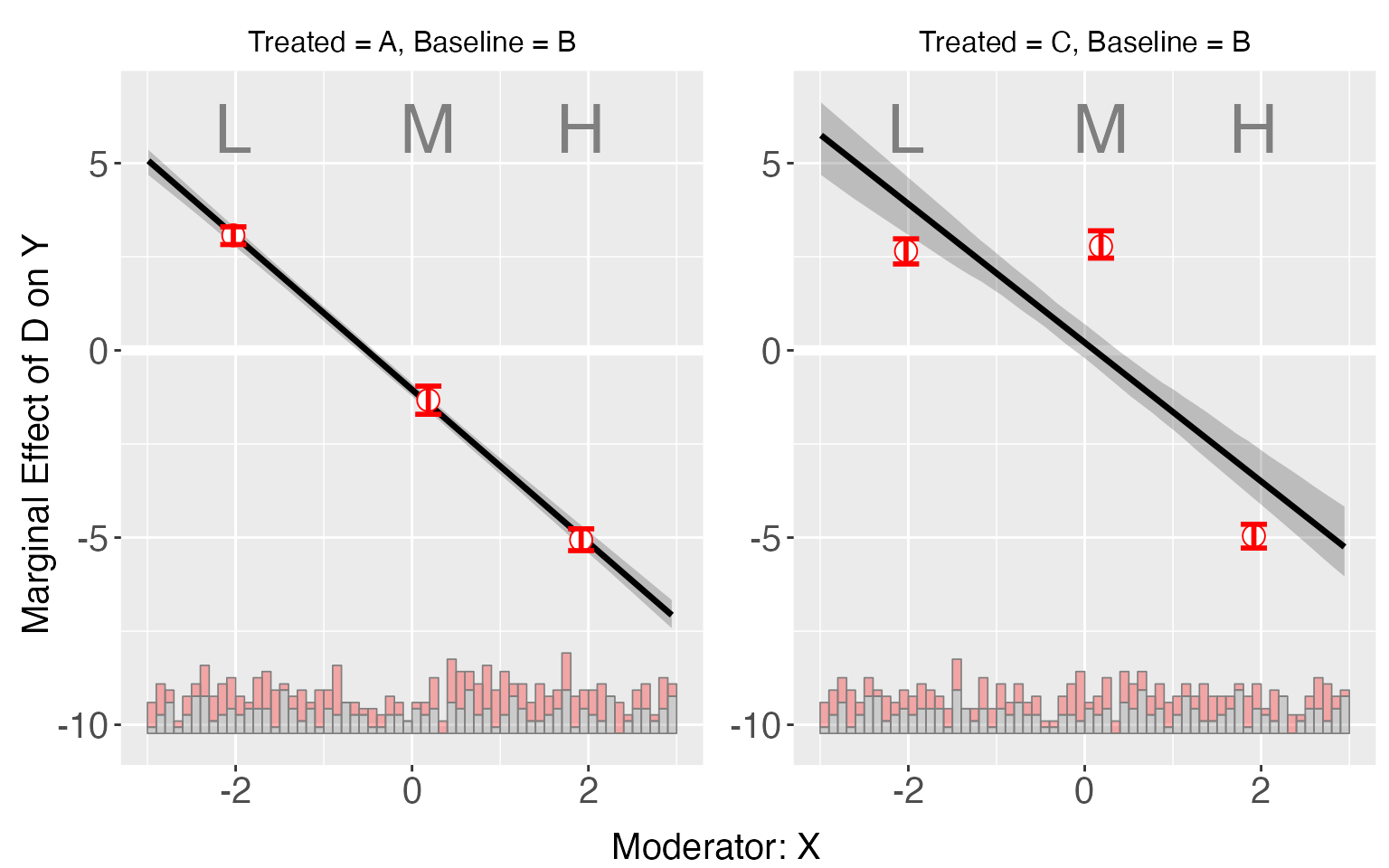
For dataset s5, because of the nonlinear interaction effects, the kernel estimator will produce CME estimates that are much closer to the effects implied by the true DGP.
s5.kernel <- interflex(Y = "Y", D = "D", X = "X",Z = c("Z1", "Z2"), data = s5,
estimator = "kernel", vartype = 'bootstrap',
nboots = 1000, parallel = TRUE, cores = 31)
s5.kernel$figure
plot(s5.kernel, pool = TRUE)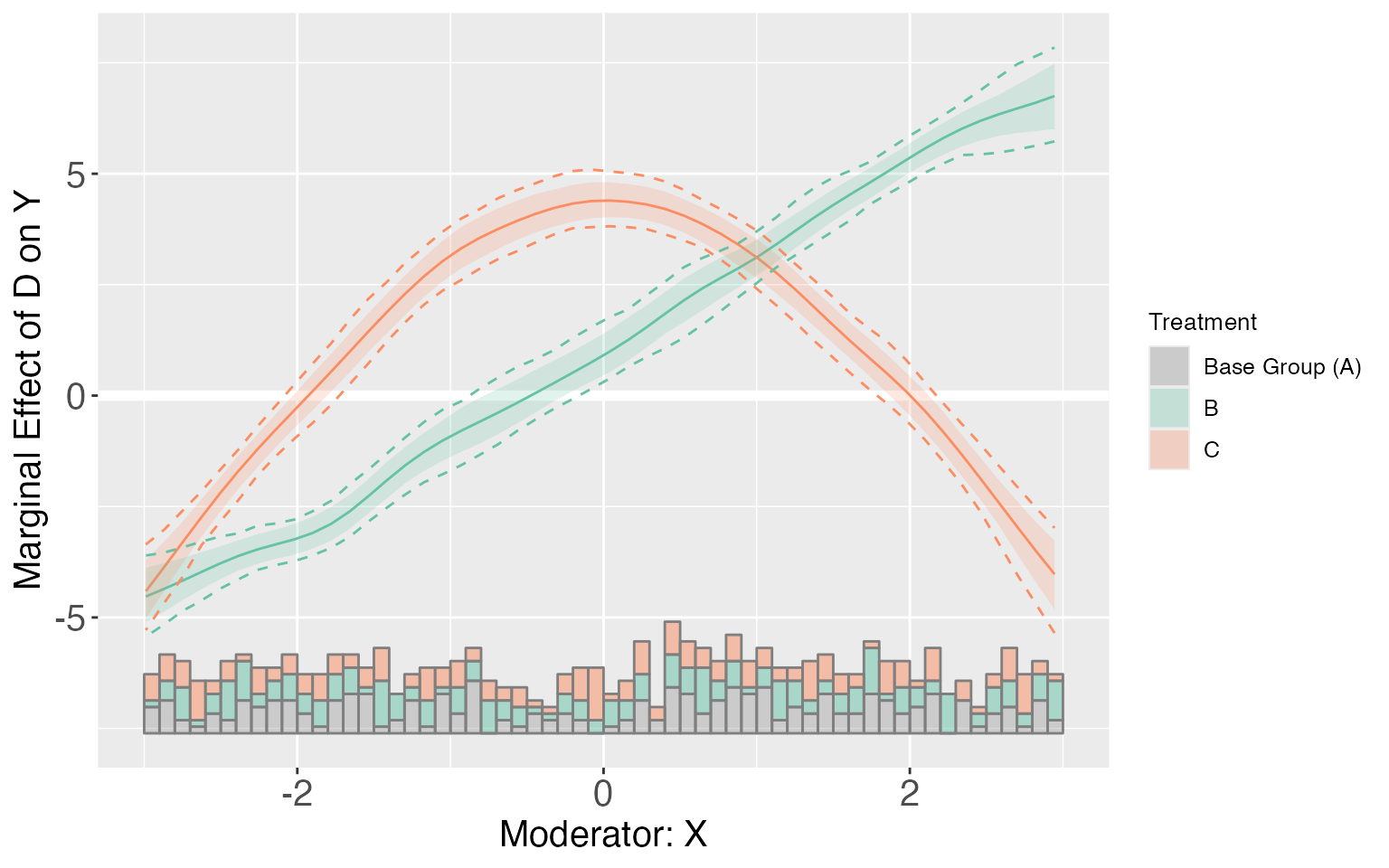
Predicted outcomes
Starting from v.1.1.0, interflex allows users to estimate and visualize predicted outcomes given fixed values of the treatment and moderator based on flexible interaction models. All predictions are made by setting covariates equal to their sample means.
s5.kernel <-interflex(Y = "Y", D = "D", X = "X", Z = c("Z1", "Z2"),
data = s5, estimator = "kernel",vartype = 'bootstrap',
nboots = 1000, parallel = TRUE, cores = 31)
predict(s5.kernel)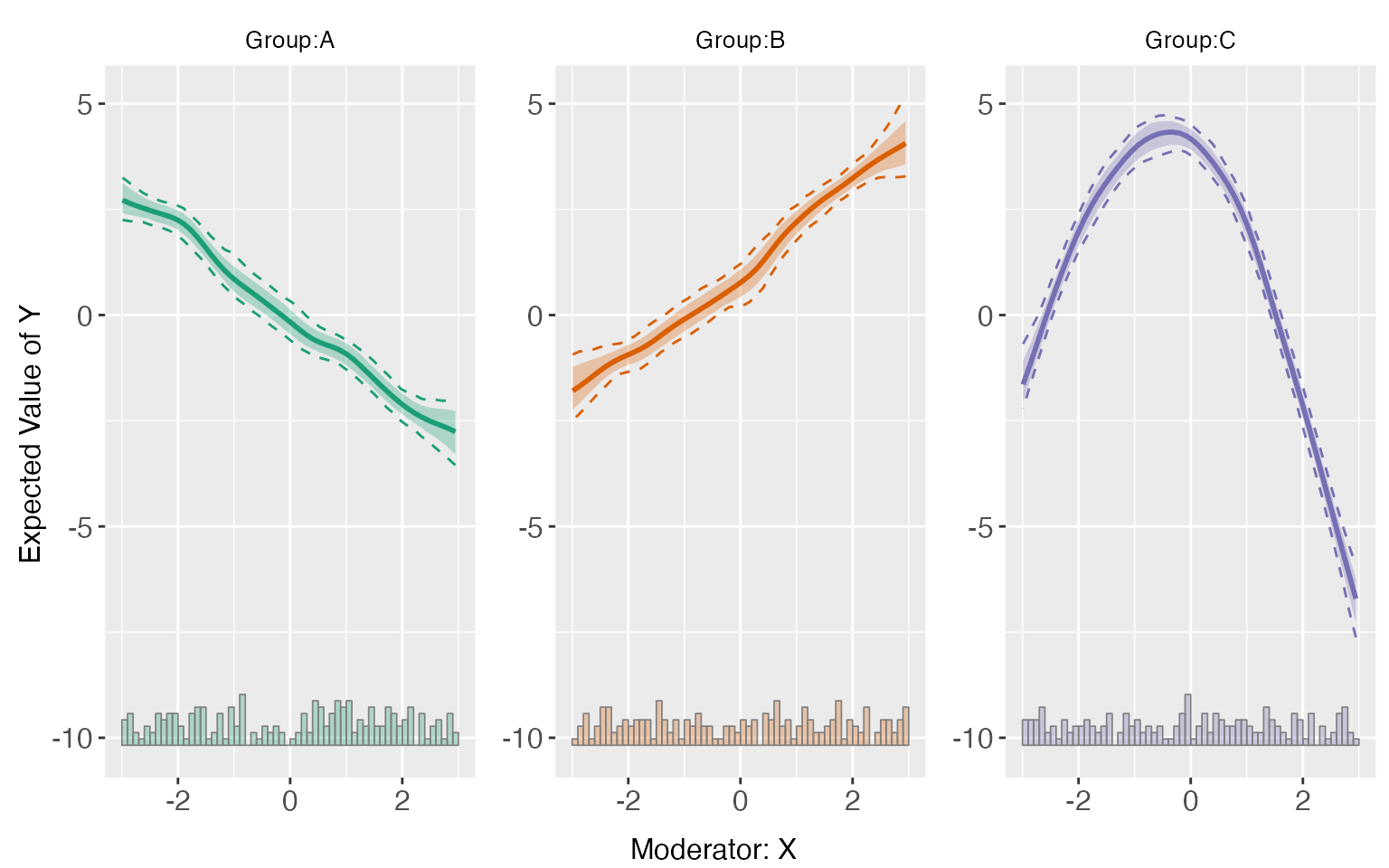
We can also set pool = TRUE to combine them.
predict(s5.kernel,order = c('A','B','C'),subtitle = c("Group A", "Group B", "Group C"),
pool = TRUE, legend.title = "Three Different Groups")
We can also use the linear model or the binning model to estimate and visualize predicted outcomes. For the latter, it is expected to see some “zigzags” at the bin boundaries in a plot:
s5.binning <-interflex(Y = "Y", D = "D", X = "X", Z = c("Z1","Z2"), data = s5,
estimator = "binning", vartype = "bootstrap", nbins = 4)
predict(s5.binning)
s5.linear <-interflex(Y = "Y", D = "D", X = "X", Z = c("Z1","Z2"), data = s5,
estimator = "linear", vcov.type = "robust")
predict(s5.linear)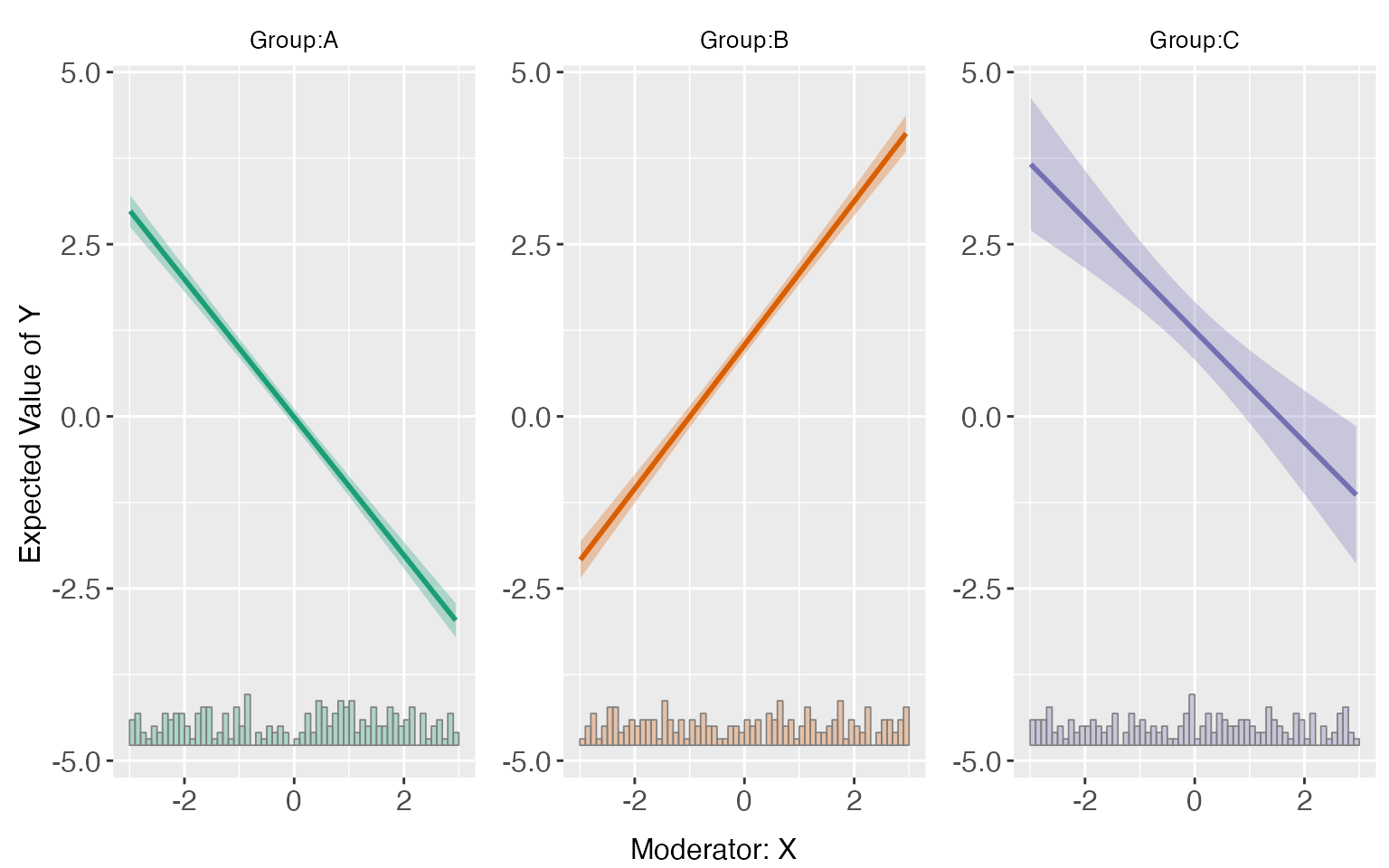
Obviously, the kernel estimator characterizes the data more better than a linear or binning estimator.
Differences in treatment effects
Starting from v.1.1.0, interflex allows users to
compare treatment effects at three specific values of the moderator
using linear or kernel model. In order to do so, users need to pass
three values into diff.values when applying linear
or kernel estimator. If not specified, this programme will by
default choose the 25,50,75 percentile of the moderator as the
diff.values.
s5.kernel <-interflex(Y = "Y", D = "D", X = "X", Z = c("Z1", "Z2"), data = s5,
estimator = "kernel", diff.values = c(-2,0,2),
vartype = "bootstrap",
nboots = 1000, parallel = TRUE, cores = 31,)
plot(s5.kernel,diff.values = c(-2,0,2))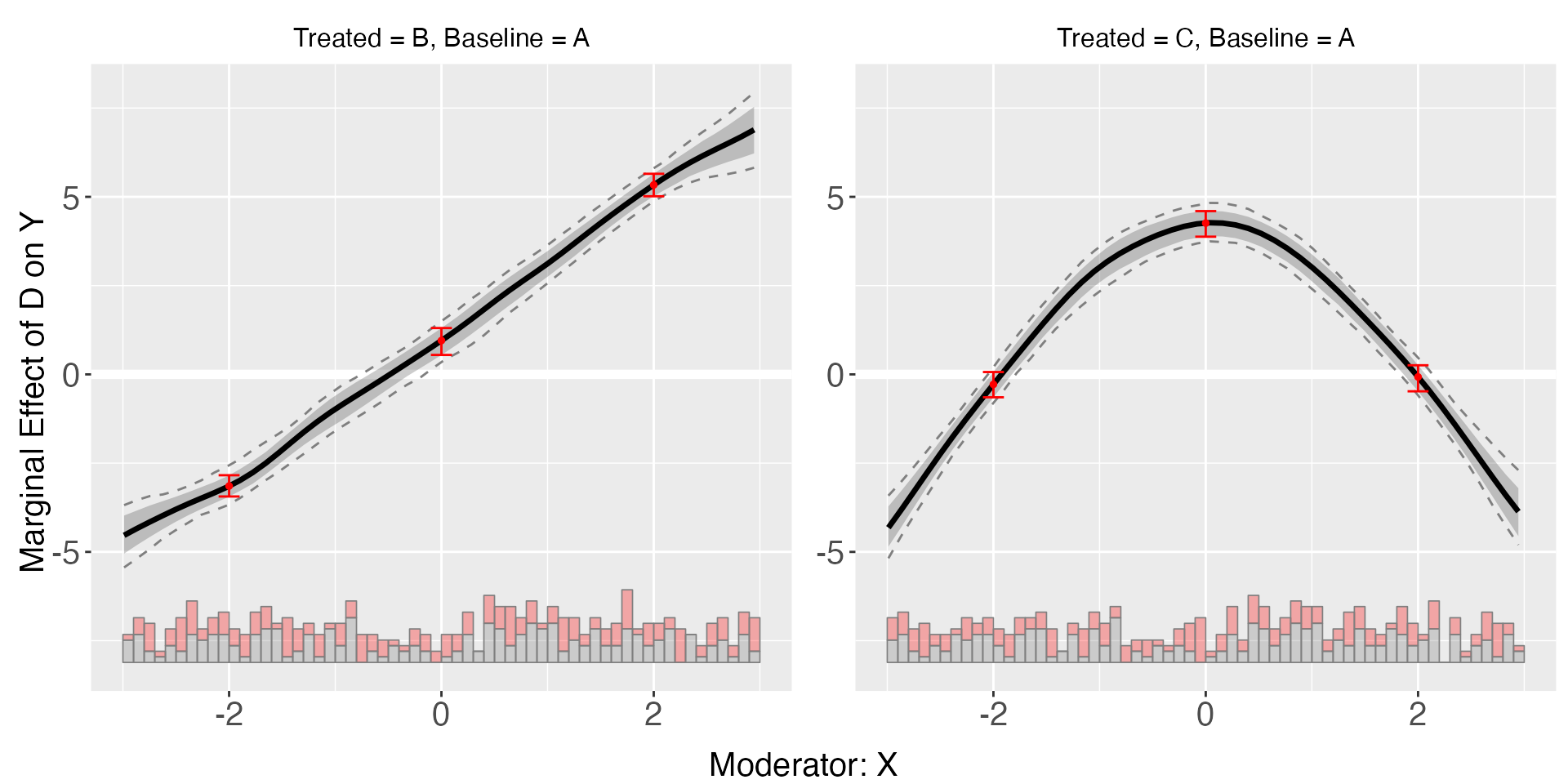
s5.kernel$diff.estimate
#> $B
#> diff.estimate sd z-value p-value lower CI(95%) upper CI(95%)
#> 0 vs -2 4.116 0.272 15.155 0.000 3.585 4.674
#> 2 vs 0 4.421 0.275 16.074 0.000 3.907 4.945
#> 2 vs -2 8.538 0.237 36.080 0.000 8.073 9.004
#>
#> $C
#> diff.estimate sd z-value p-value lower CI(95%) upper CI(95%)
#> 0 vs -2 4.611 0.270 17.066 0.000 4.103 5.152
#> 2 vs 0 -4.373 0.280 -15.590 0.000 -4.910 -3.813
#> 2 vs -2 0.238 0.284 0.839 0.402 -0.267 0.832
s5.linear <-interflex(Y = "Y", D = "D", X = "X", Z = c("Z1","Z2"), data = s5,
estimator = "linear", diff.values = c(-2,0,2),
vartype = "bootstrap")
plot(s5.linear,diff.values = c(-2,0,2))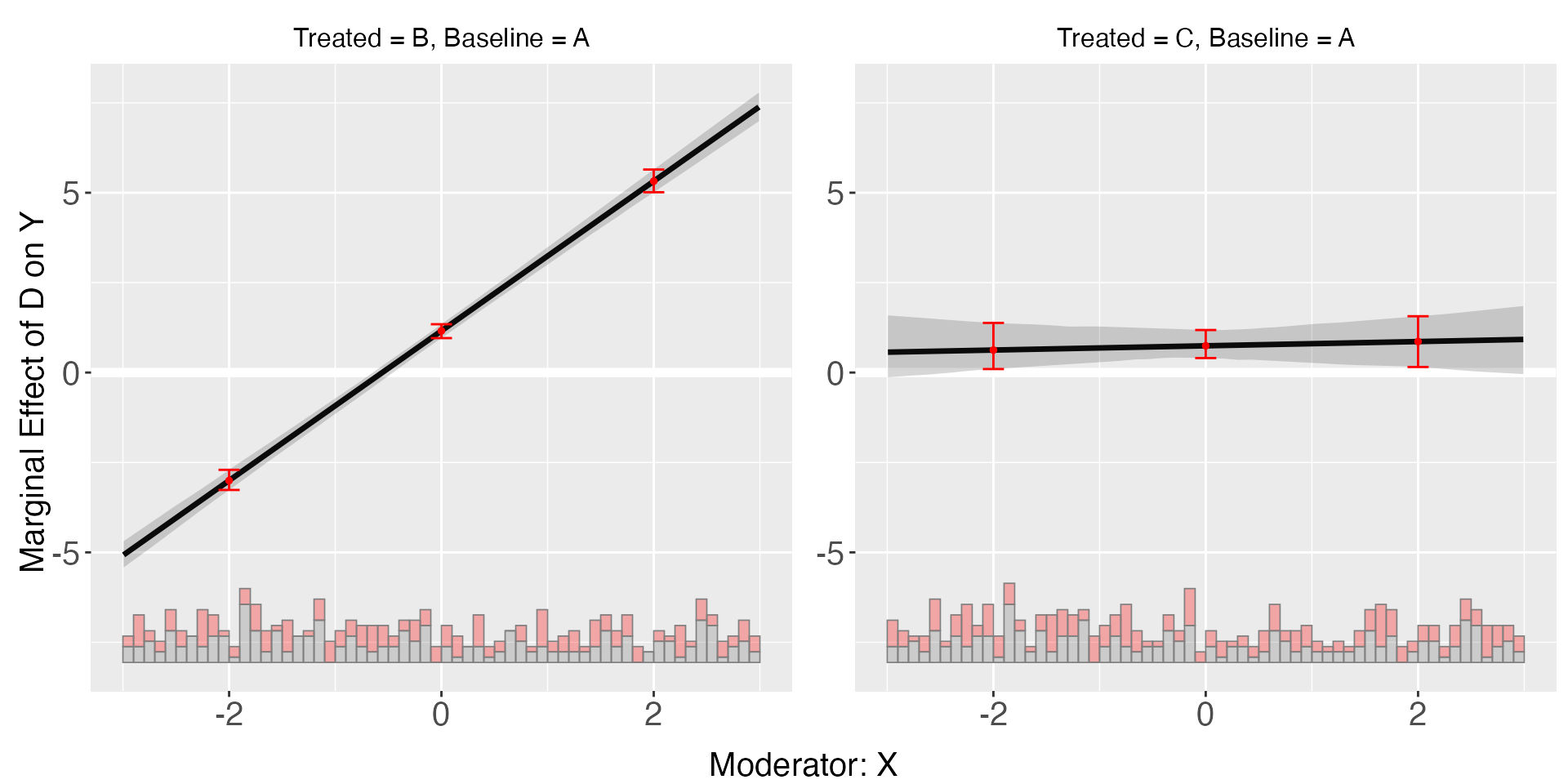
s5.linear$diff.estimate
#> $B
#> diff.estimate sd z-value p-value lower CI(95%) upper CI(95%)
#> 0 vs -2 4.090 0.096 42.534 0.000 3.882 4.253
#> 2 vs 0 4.090 0.096 42.534 0.000 3.882 4.253
#> 2 vs -2 8.179 0.192 42.534 0.000 7.764 8.505
#>
#> $C
#> diff.estimate sd z-value p-value lower CI(95%) upper CI(95%)
#> 0 vs -2 0.384 0.363 1.058 0.290 -0.251 1.071
#> 2 vs 0 0.384 0.363 1.058 0.290 -0.251 1.071
#> 2 vs -2 0.769 0.727 1.058 0.290 -0.501 2.141Starting from v.1.1.1, interflex allows users to
compare treatment effects at two or three specific values of the
moderator using CME and vcov matrix derived from linear/kernel
estimation. Based on GAM model(relies on mgcv package),
users can approximate treatment effects and their variance using smooth
functions without re-estimating the model, hence saving time. In order
to do so, users need to pass the output of interflex
into t.test and then specify the values of interest of
the moderator. As it is an approximation, the results here are expected
to have a little deviance from the output we got by directly specifying
diff.values when applying inter.linear or
inter.kernel. We also limit the elements passed in
diff.values between the minimum and the maximum of the moderator to
avoid potential extrapolation bias.
inter.test(s5.kernel,diff.values=c(-2,0,2))
#> $B
#> diff.estimate sd z-value p-value lower CI(95%) upper CI(95%)
#> 0 vs -2 4.106 0.271 15.141 0.000 3.574 4.637
#> 2 vs 0 4.425 0.275 16.110 0.000 3.887 4.964
#> 2 vs -2 8.531 0.236 36.074 0.000 8.068 8.995
#>
#> $C
#> diff.estimate sd z-value p-value lower CI(95%) upper CI(95%)
#> 0 vs -2 4.605 0.269 17.094 0.000 4.077 5.132
#> 2 vs 0 -4.372 0.280 -15.625 0.000 -4.921 -3.824
#> 2 vs -2 0.232 0.283 0.820 0.412 -0.322 0.786
inter.test(s5.linear,diff.values=c(-2,0,2))
#> $B
#> diff.estimate sd z-value p-value lower CI(95%) upper CI(95%)
#> 0 vs -2 4.090 0.096 42.534 0.000 3.901 4.278
#> 2 vs 0 4.090 0.096 42.534 0.000 3.901 4.278
#> 2 vs -2 8.179 0.192 42.534 0.000 7.802 8.556
#>
#> $C
#> diff.estimate sd z-value p-value lower CI(95%) upper CI(95%)
#> 0 vs -2 0.384 0.363 1.058 0.290 -0.328 1.097
#> 2 vs 0 0.384 0.363 1.058 0.290 -0.328 1.097
#> 2 vs -2 0.769 0.727 1.058 0.290 -0.656 2.193We can also set percentile = TRUE to use the percentile
scale rather than the real scale of the moderator.
inter.test(s5.kernel,diff.values=c(0.25,0.5,0.75),percentile=TRUE)
#> $B
#> diff.estimate sd z-value p-value lower CI(95%) upper CI(95%)
#> 50% vs 25% 3.046 0.273 11.154 0.000 2.510 3.581
#> 75% vs 50% 3.307 0.280 11.802 0.000 2.757 3.856
#> 75% vs 25% 6.352 0.260 24.399 0.000 5.842 6.862
#>
#> $C
#> diff.estimate sd z-value p-value lower CI(95%) upper CI(95%)
#> 50% vs 25% 2.792 0.269 10.372 0.000 2.265 3.320
#> 75% vs 50% -2.627 0.270 -9.722 0.000 -3.156 -2.097
#> 75% vs 25% 0.166 0.285 0.581 0.561 -0.393 0.725