# load libraries and data
library(ggplot2)
library(panelView)
library(fect)
data(fect)
# Define variables
y <- "general_sharetotal_A_all" # Outcome: Share of Asian contributions
d <- "cand_A_all" # Treatment: Presence of an Asian candidate
controls <- c("cand_H_all", "cand_B_all") # Controls: Presence of Hispanic and Blank candidates
unit <- "district_final" # Unit variable: District
time <- "cycle" # Time variable: Election cycle
clust <- "district_final" # Clustering variable: District3 Fect Plot Options
In this chapter, we explore various visualization options avialable in the fect package using data from Grumbach and Sahn (2020). This chapter is authored by Jinwen Wu and Yiqing Xu.
plot.fect is an S3 method that offers various options for customizing data and results visualization. Below is a brief summary of the most commonly used options.
-
Starting Period:
-
start0: IfTRUE, shifts the time axis so that treatment begins at Period 0 instead of Period 1.
-
-
Confidence Intervals:
-
plot.ci: Options include"none","90", or"95"to hide confidence intervals or display 90% or 95% confidence intervals.
-
-
Axis and Legend Customization:
-
xlim/ylim: Set the x- and y-axis ranges.
-
xlab/ylab: Customize axis labels.
-
xbreaks/ybreaks: Specify tick marks.
-
xangle/yangle: Adjust the rotation angle of axis text.
-
legend.pos,legend.nrow,legend.labs: Control legend placement, number of rows, and labels.
-
-
Theme and Text:
-
theme.bw: IfTRUE, applies a black-and-white theme.
-
cex.main,cex.axis,cex.lab,cex.text: Adjust text sizes for the title, tick labels, axis labels, and annotations.
-
-
Lines and Bounds:
-
line.color/line.width: Define the color and width of main lines.
-
lcolor/lwidth: Set the color and width for the horizontal zero line.
-
While these customization options are demonstrated using the default gap plot, they can be applied universally, with only a few exceptions.
3.1 Load Data
As explained in the previous chapter, Grumbach and Sahn (2020) examines the mobilizing effect of minority candidates on coethnic support in U.S. congressional elections. The treatment variable indicates the presence of an Asian candidate, and the outcome variable represents the proportion of general election contributions from Asian donors.
First, we load the required packages. The dataset, gs2020, is included with the fect package and can be loaded using data(fect). We also specify the treatment, outcome, controls (covariates), unit and time indices, and level of clustering.
3.2 Gap Plot
To create the gap plot, also known as the event study plot, we first apply fect, the fixed effects counterfactual estimator. For details, see Chapter 2.
out <- fect(Y = y, D = d, X = controls, index = c(unit, time),
data = gs2020, method = "fe", force = "two-way",
se = TRUE, parallel = TRUE, nboots = 100)
#> Some units are totally removed after drop missing values of the outcome or covariates.
#> Parallel computing ...
#> Bootstrapping for uncertainties ...
#> 100 runsAfter running the model, we can plot the dynamic treatment effects over (relative) time, including confidence intervals if se = TRUE is specified in the estimation. Note that type = "gap" is the default option, so we omit it here.
plot(out)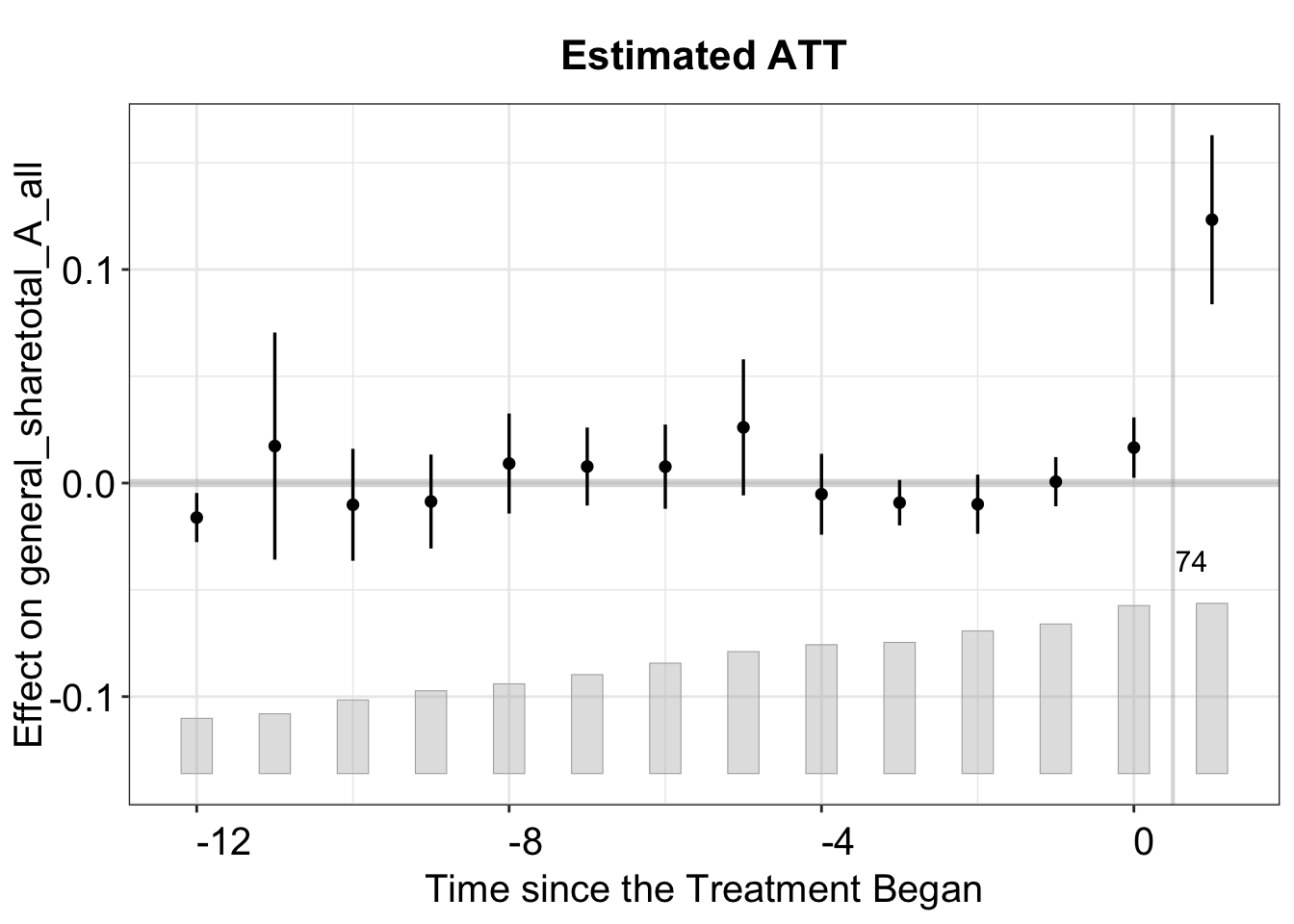
3.2.1 Starting Period
By default, the first post-treatment period is set to 0, and the last pre-treatment period is set to -1. However, some researchers prefer to designate the former as 1 and the latter as 0. To achieve this, set start0 = TRUE.
plot(out, start0 = TRUE, # Shift time so treatment begins at 0
main = "Custom Starting Period")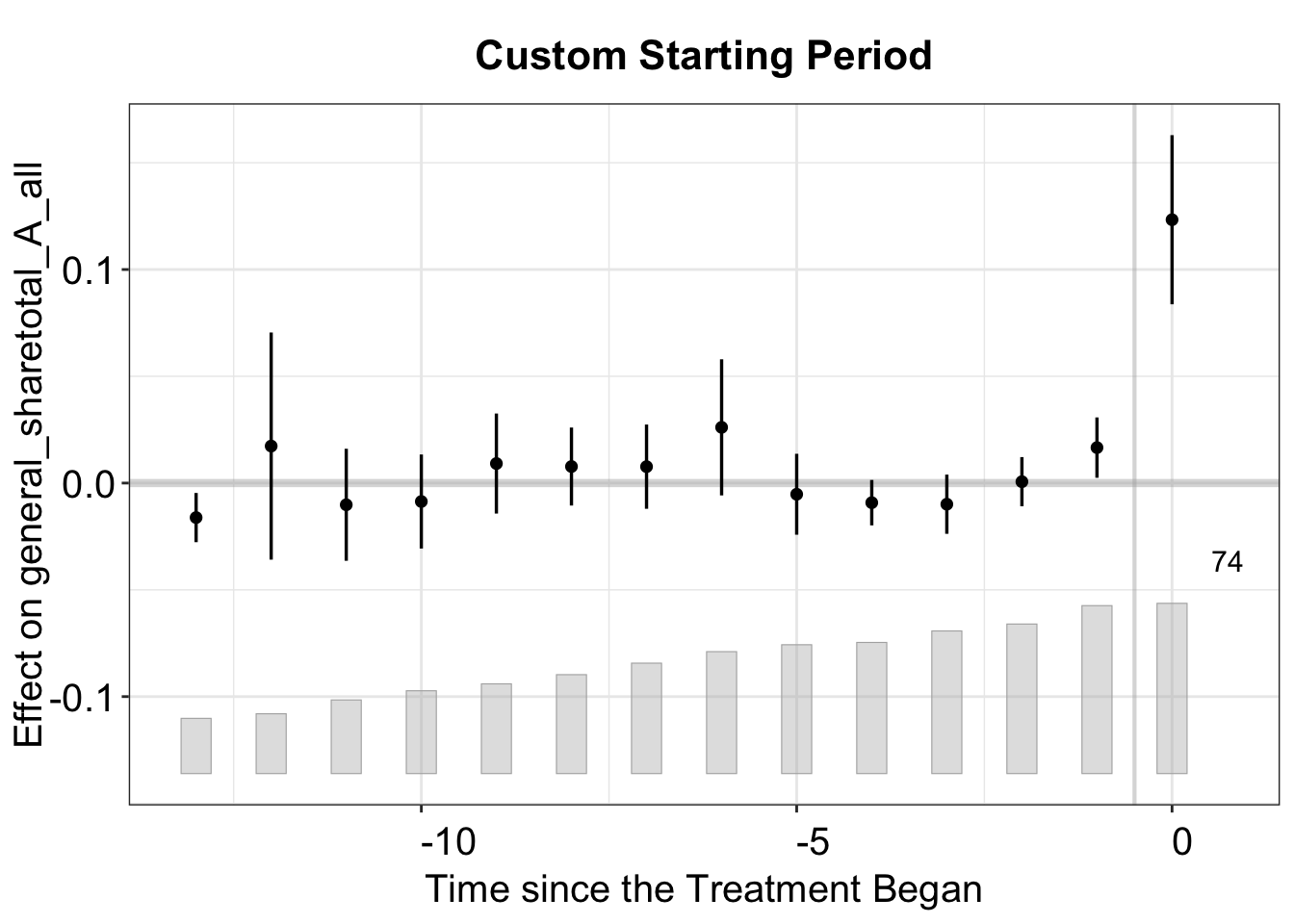
3.2.2 Axis and Legend
Below, we customize the x- and y-axis ranges, labels, tick breaks, and legend. The x-axis labels are rotated for clarity. Moreover, by setting xlim = c(-10, 1), the x-axis is restricted to time periods -8 to 1, with the treatment shifted to begin at period 0 instead of period 1.
plot(out,
xlim = c(-10, 1), # only show time periods -8 to 1
ylim = c(-0.15, 0.30), # set y-range
xlab = "Custom Time Axis", # x-axis label
ylab = "Estimated ATT", # y-axis label
xangle = 90, # rotate x-axis labels by 90°
xbreaks = seq(-10, 1, by = 2),
# Label x-axis from -12 to 1 with a break of 2
main = "Axis and Legend Customization")
#> Scale for x is already present.
#> Adding another scale for x, which will replace the existing scale.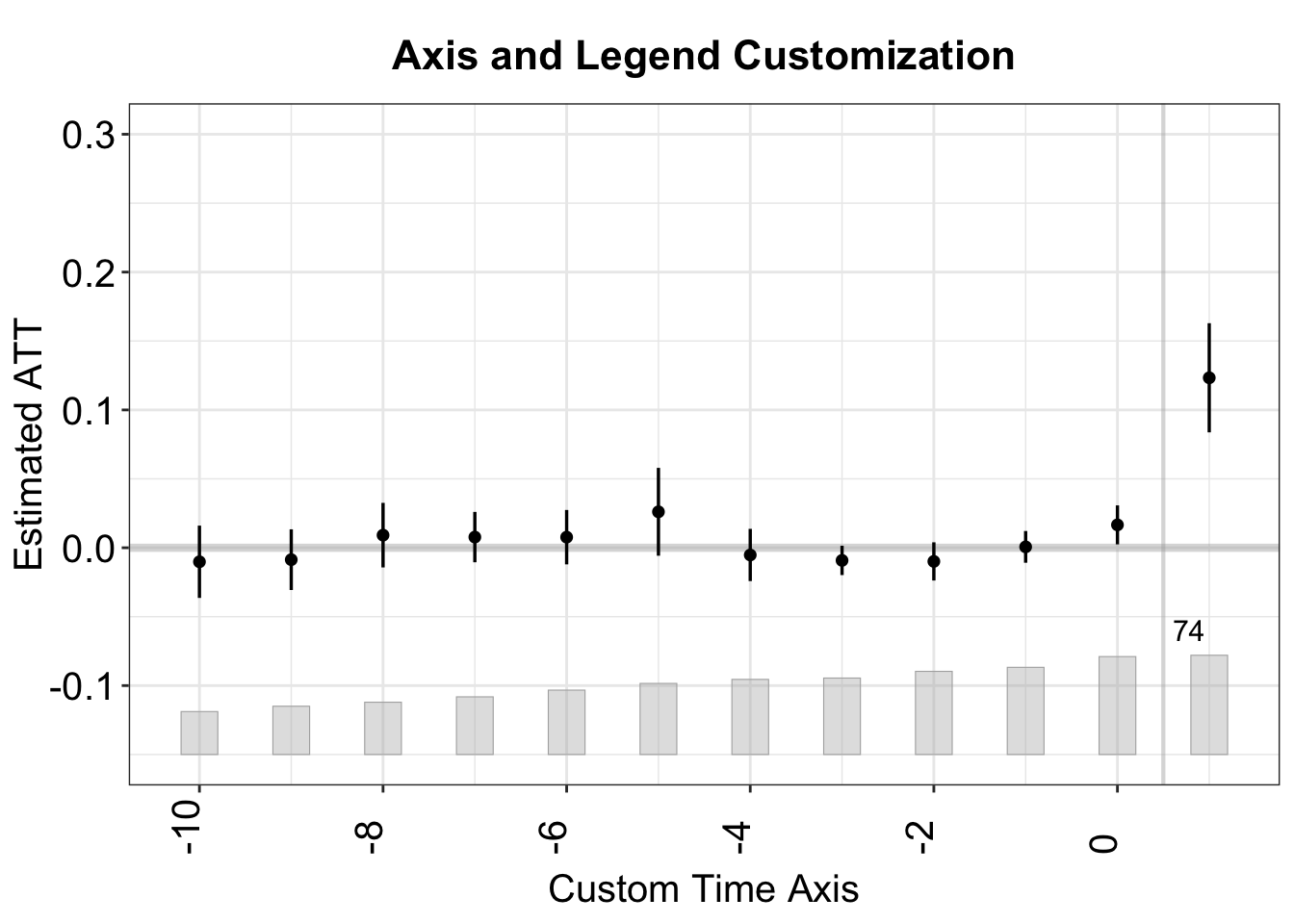
3.2.3 Confidence Intervals
Below, we plot the treatment effect with 90% confidence intervals instead of 95%.
plot(out, plot.ci = "0.9",
main = "90% confidence intervals")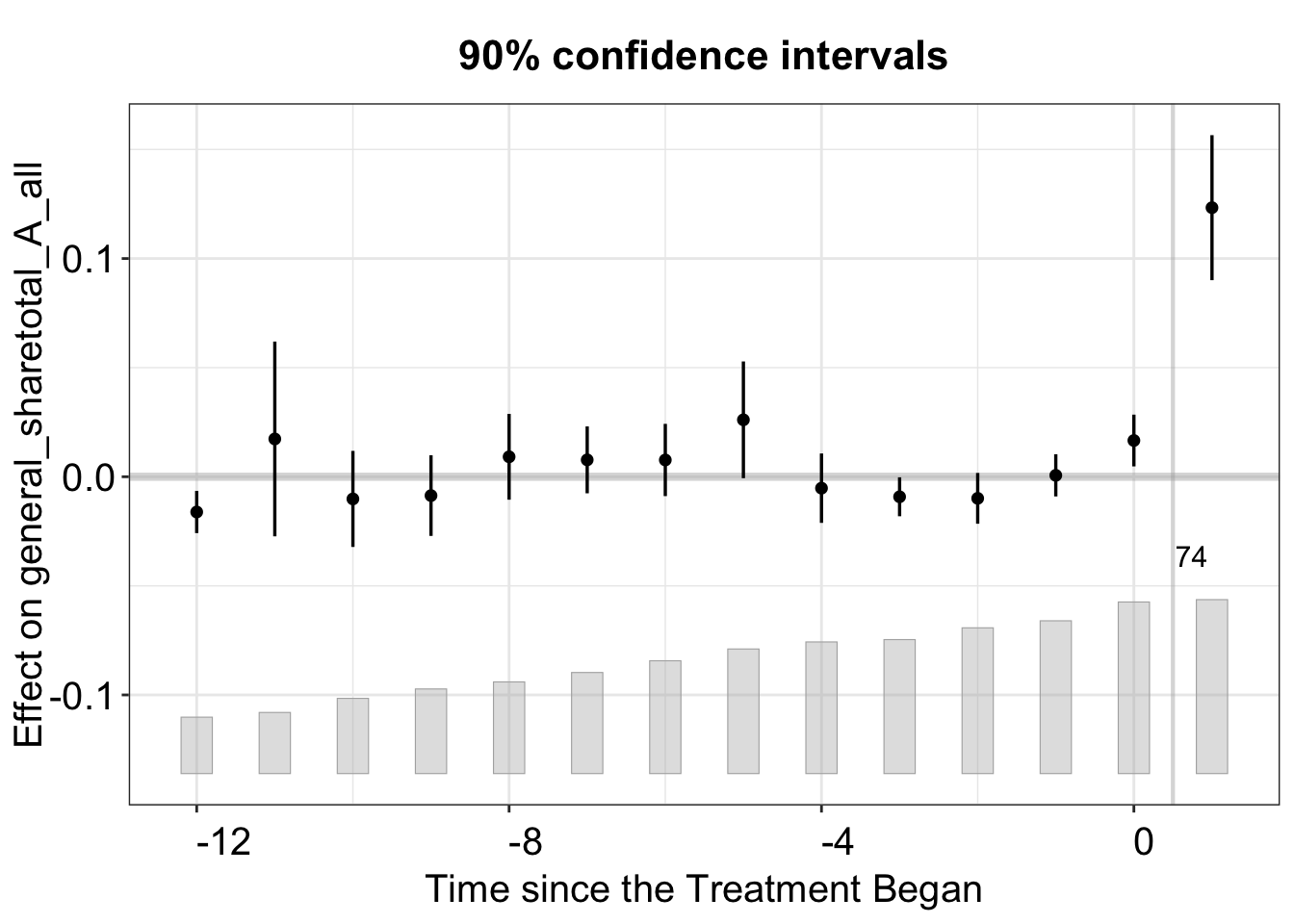
3.2.4 Text and Theme
This plot adjusts text sizes with a series of cex options.
plot(out,
ylim = c(-0.15, 0.3), # set yrange
theme.bw = FALSE, # Change the color theme
cex.main = 1.25, # Scale for the main title
cex.axis = 1.2, # Axis tick label size
cex.lab = 1.2, # Axis label size
cex.legend = 1, # Legend text size
cex.text = 1.2, # Annotation text size
main = "Text and Theme Customization")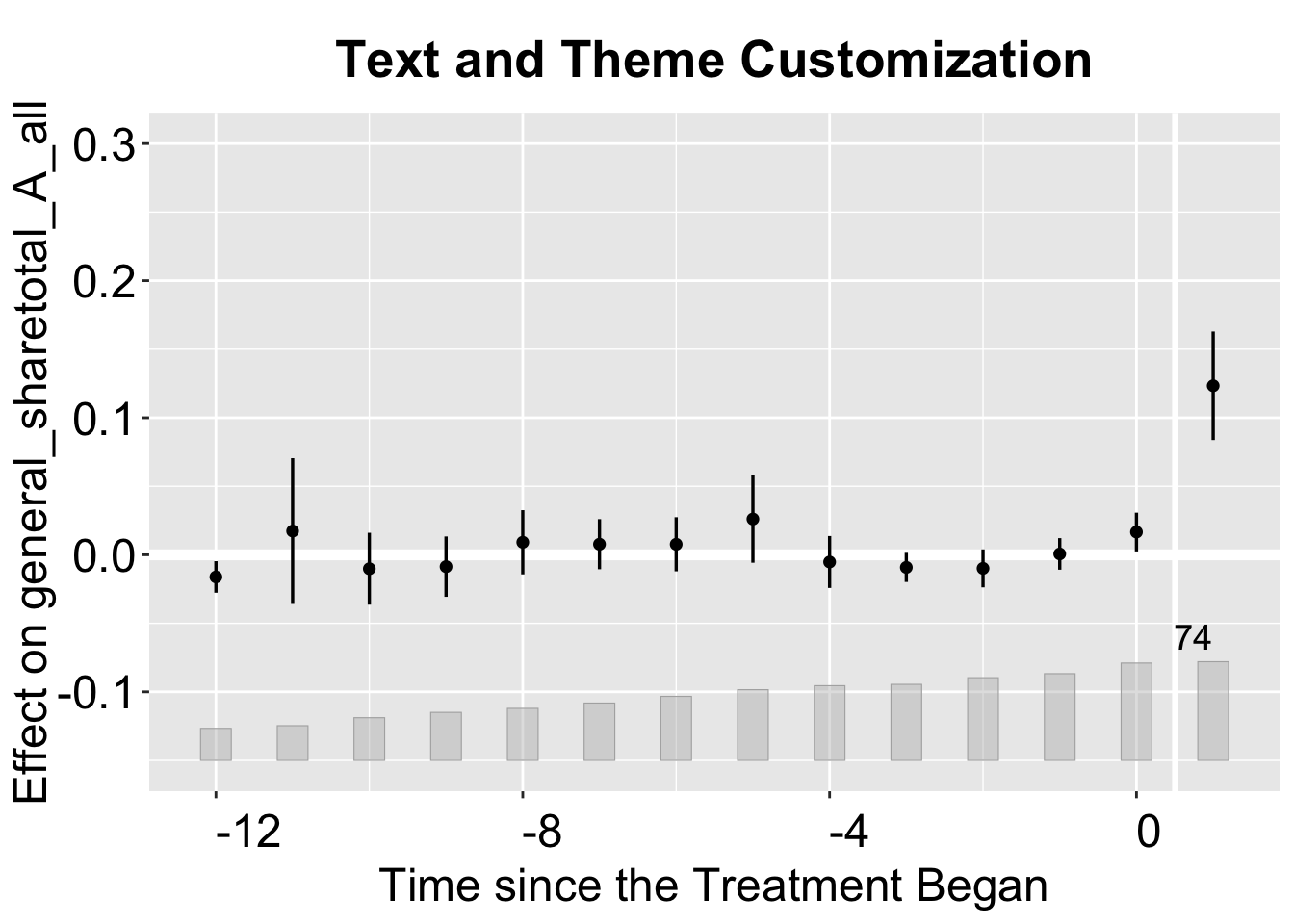
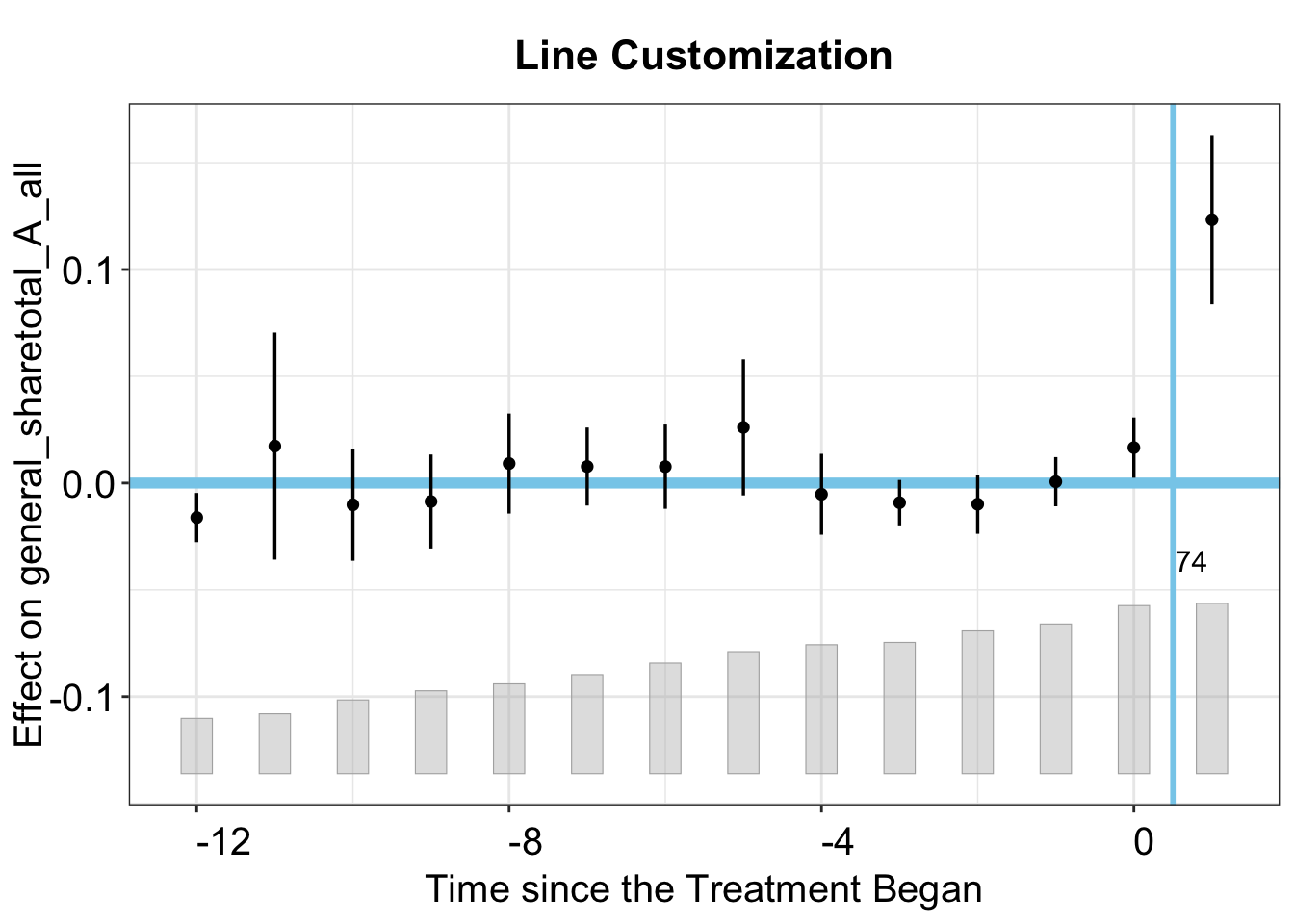
3.2.5 Line Customization
Here, we demonstrate how to change main and the horizontal reference lines.
3.3 Pretrend Tests
We can conduct several tests to shed light on (not directly test) the parallel trends (PT) assumption, including the equivalence test and the placebo test. For details, see Chapter 2 or Liu, Wang, and Xu (2024).
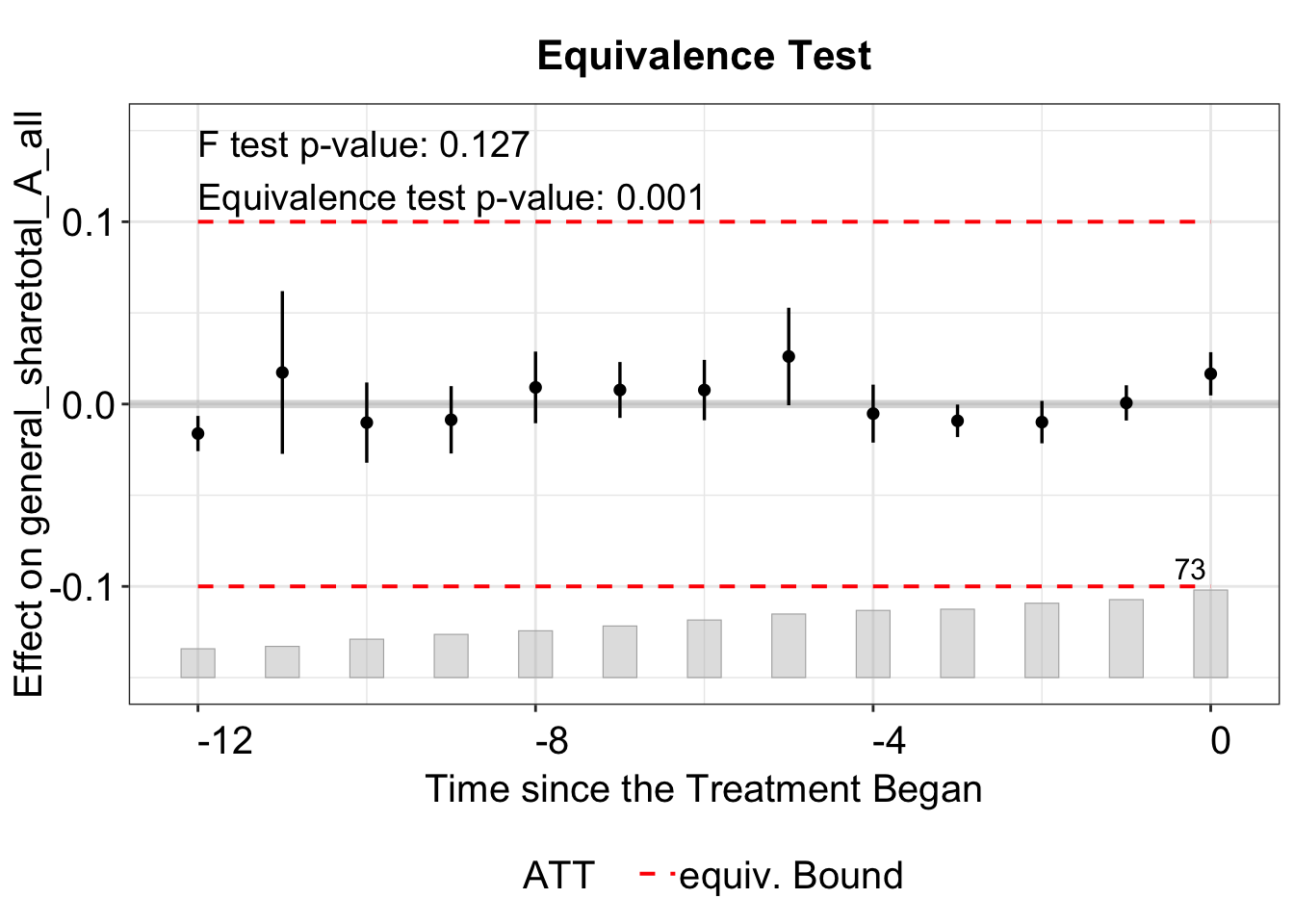
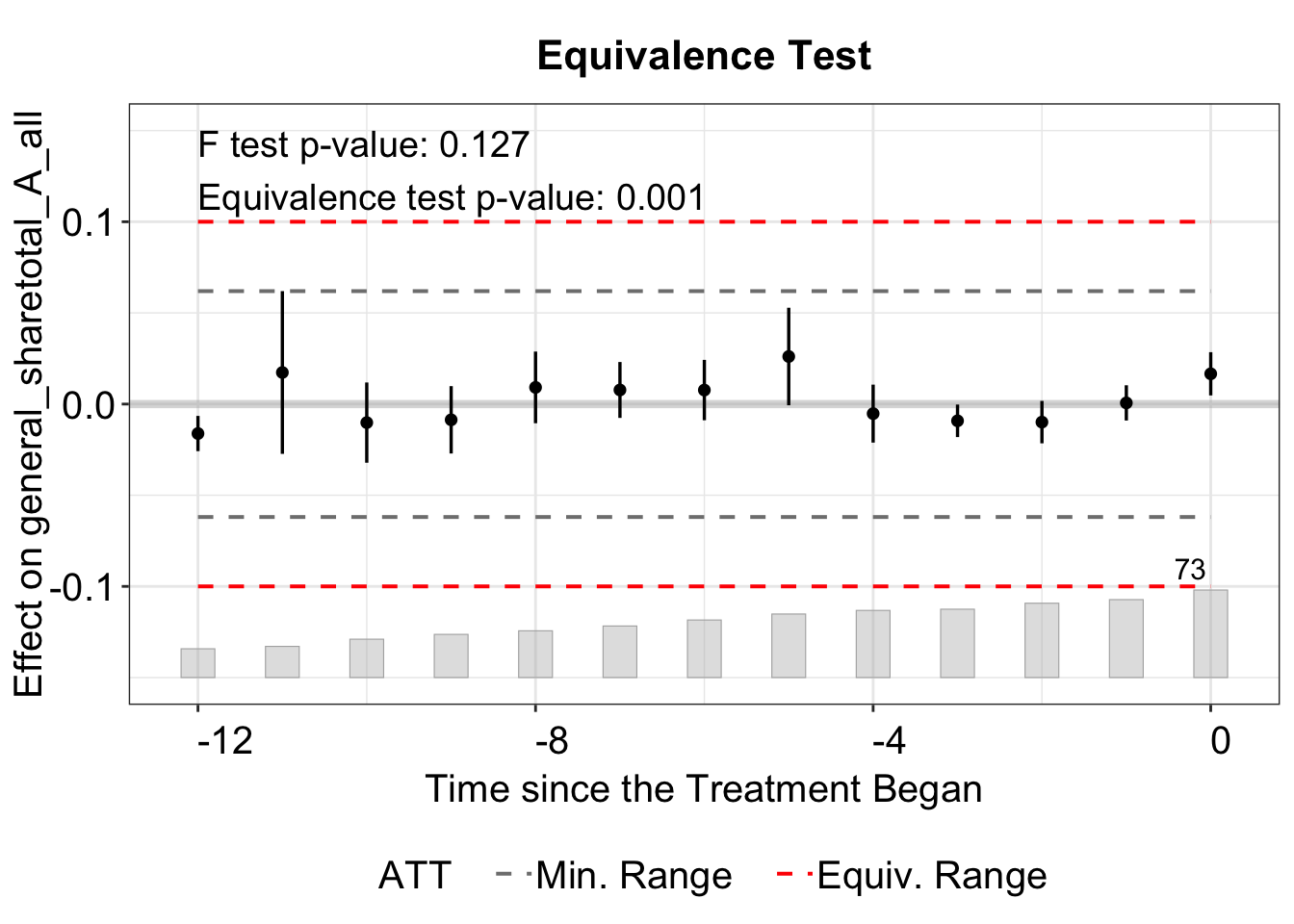
3.3.1 Equivalence Test
In the equivalence plot (type = "equiv"), the equivalence bound is defined by the two-one-sided test (TOST) threshold. For example, in the plot below, the bound is set by tost.threshold = 0.1, with lines at -0.1 and 0.1. This threshold should be set based on the magnitude of the ATT or the standard deviation of the outcome (or residualized outcome).
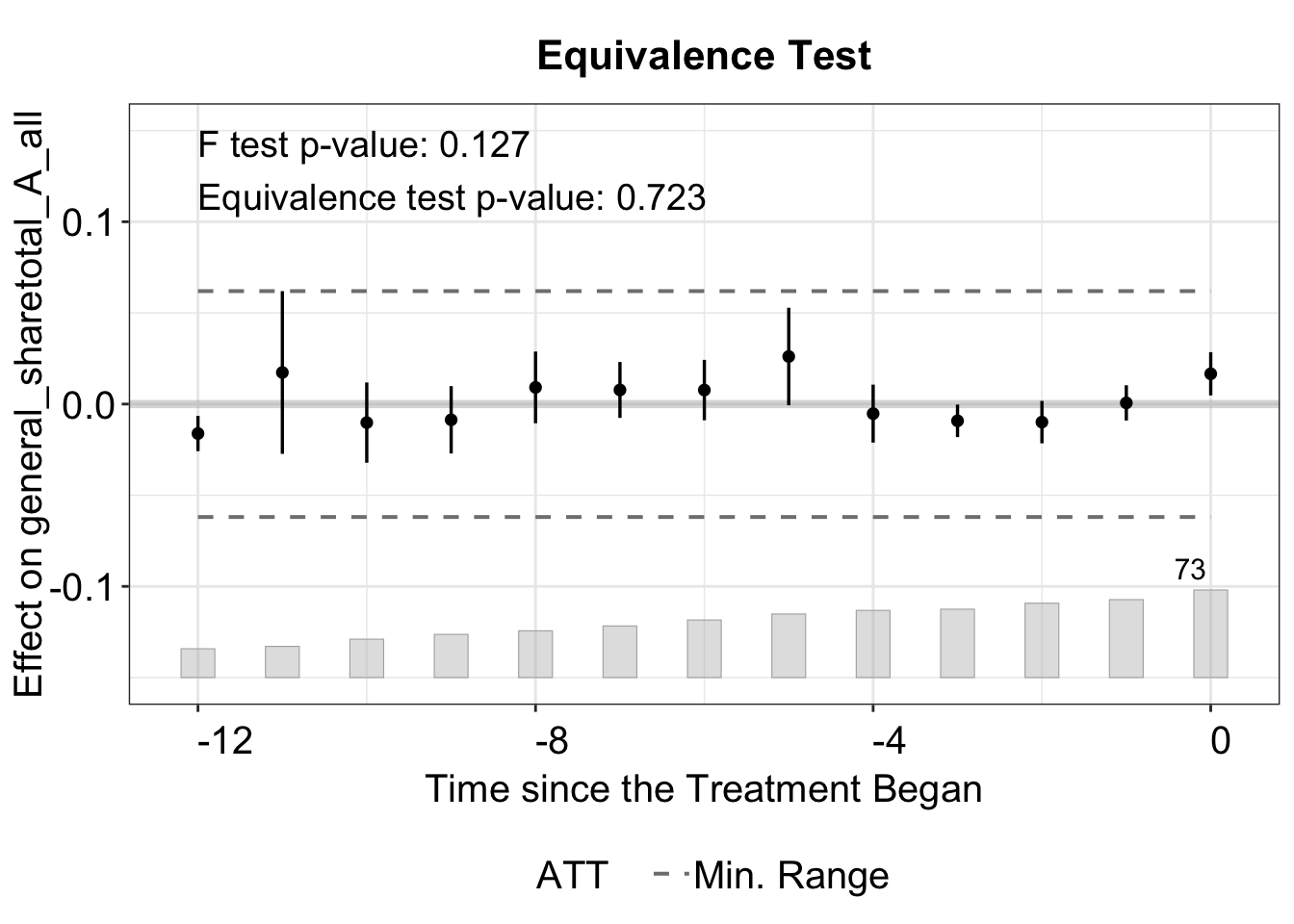
The bound option has four choices: "none", "min", "equiv", or "both". When set to "none", no bound is displayed.
The "min" displays the minimum range bound based on the maximum absolute pre‐treatment residual (e.g., if the largest pre-treatment estimate is 0.3, lines at -0.3 and 0.3).
We can plot both the minimum range and the equivalence bound with bound = "both", which is also the default option.
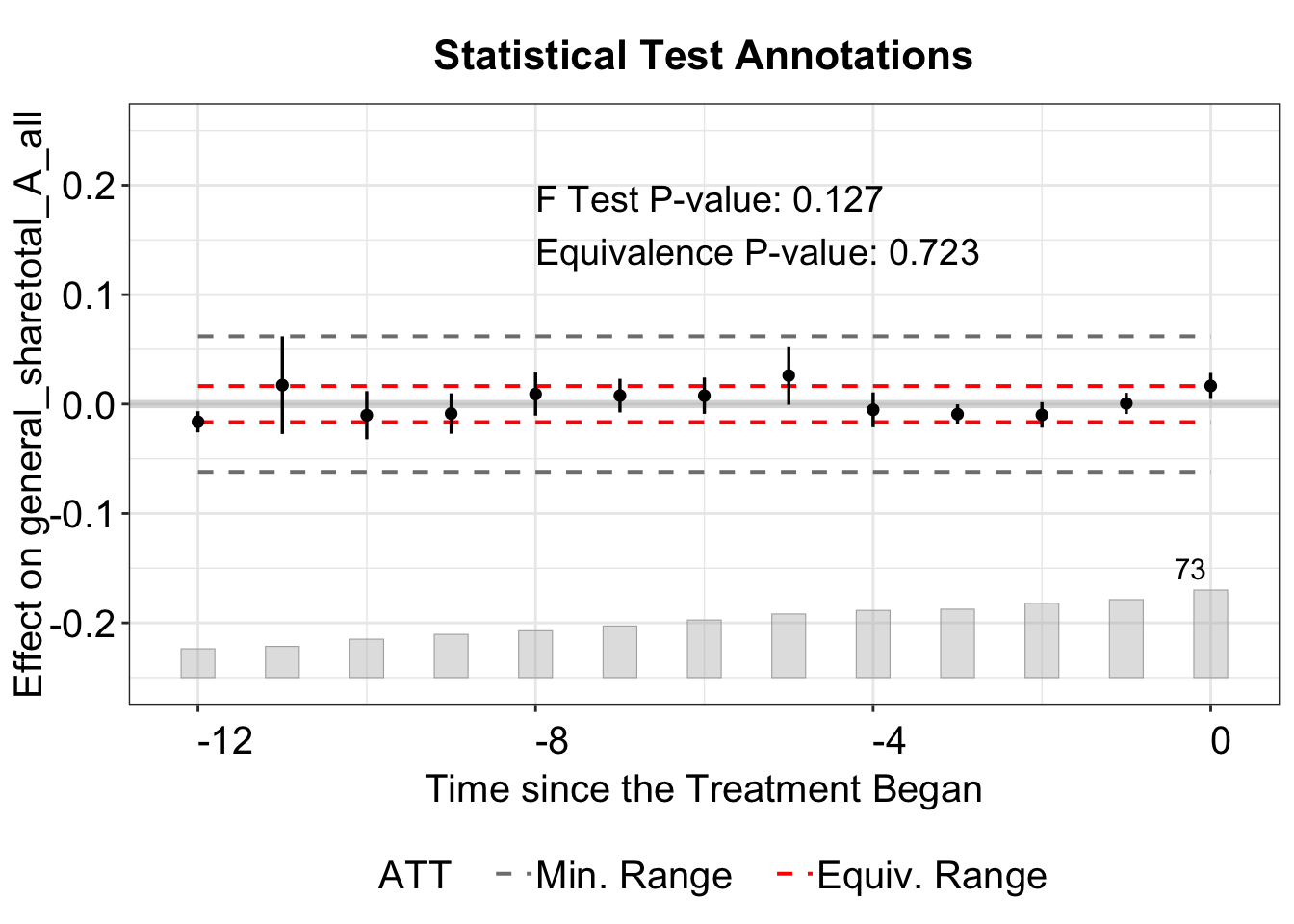
We use the stats argument to select which results to display, label them with stats.labs, and position the legend with stats.pos. Setting show.stats = FALSE hides the test results entirely.
3.3.2 Placebo Test
A placebo test evaluates whether the “fake” ATT is statistically distinguishable in a placebo period. It artificially assigns treatment during placebo periods and estimates the “placebo effect” in those periods.
Therefore, the model must be re-run. Below, we set placebo.period = c(-2, 0), specifying the pre-treatment periods used for the placebo test.
out_fe_placebo <- fect(Y = y, D = d, X = controls, data = gs2020,
index = c(unit, time), force = "two-way",
method = "fe", CV = FALSE, parallel = TRUE,
se = TRUE, nboots = 1000, placeboTest = TRUE,
placebo.period = c(-2, 0))
#> Some units are totally removed after drop missing values of the outcome or covariates.
#> Parallel computing ...
#> Bootstrapping for uncertainties ...
#> 1000 runs
plot(out_fe_placebo)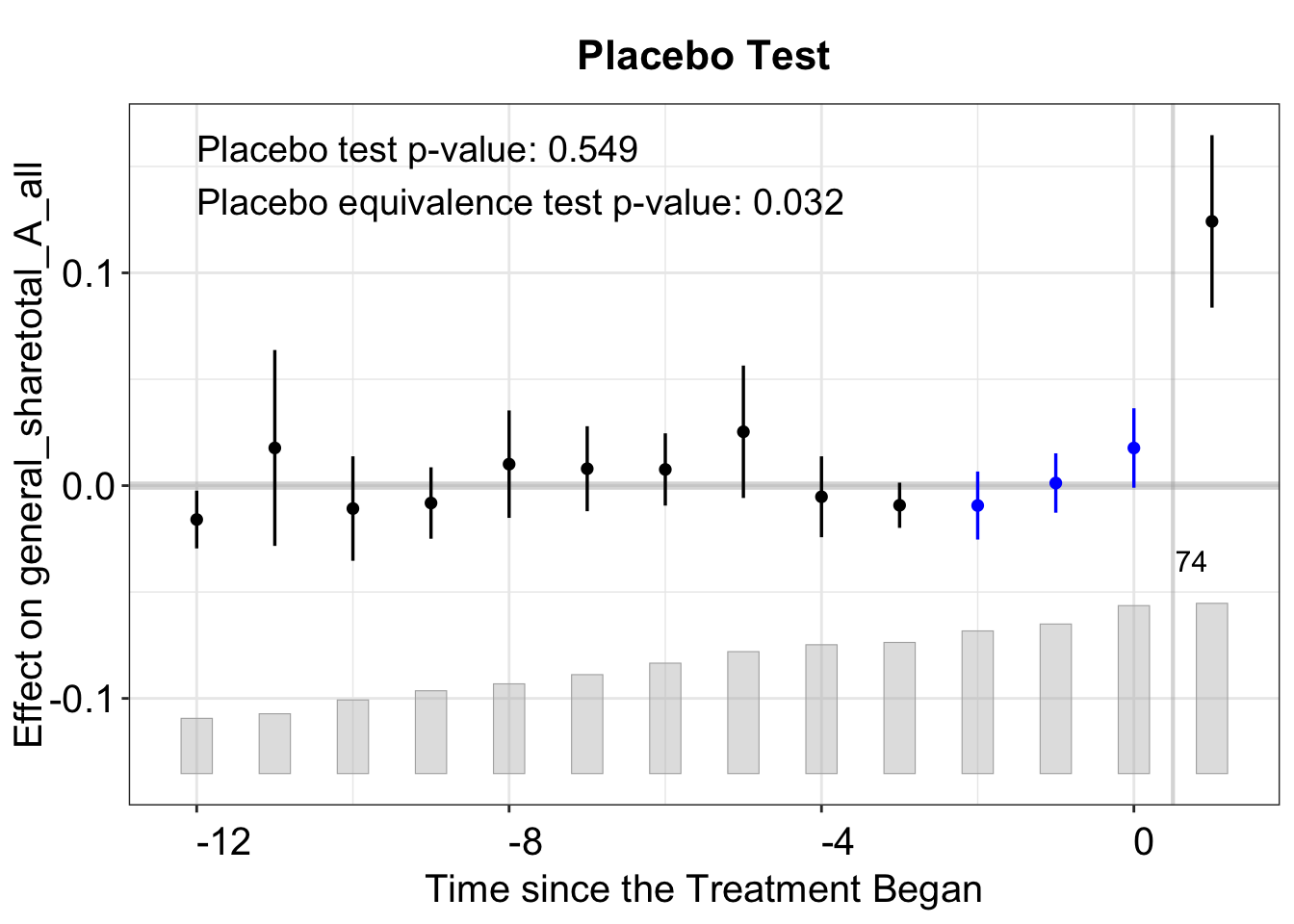
The plot shows the estimated “effects” in the pre-treatment periods (placebo effects). The blue lines in the pre-treatment period suggest that we do not observe significant effects of the treatment in the pre-periods.
3.4 Carryover Effects
One type of plot rarely seen in the empirical literature is how the difference between treatment and control groups evolves after treatment ends. We call it the "exit" plot, where the x-axis represents time relative to treatment exit. In contrast, the "gap" plot focuses on treatment entry. The "exit" plot is essential for assessing potential carryover effects.
plot(out, type = "exit")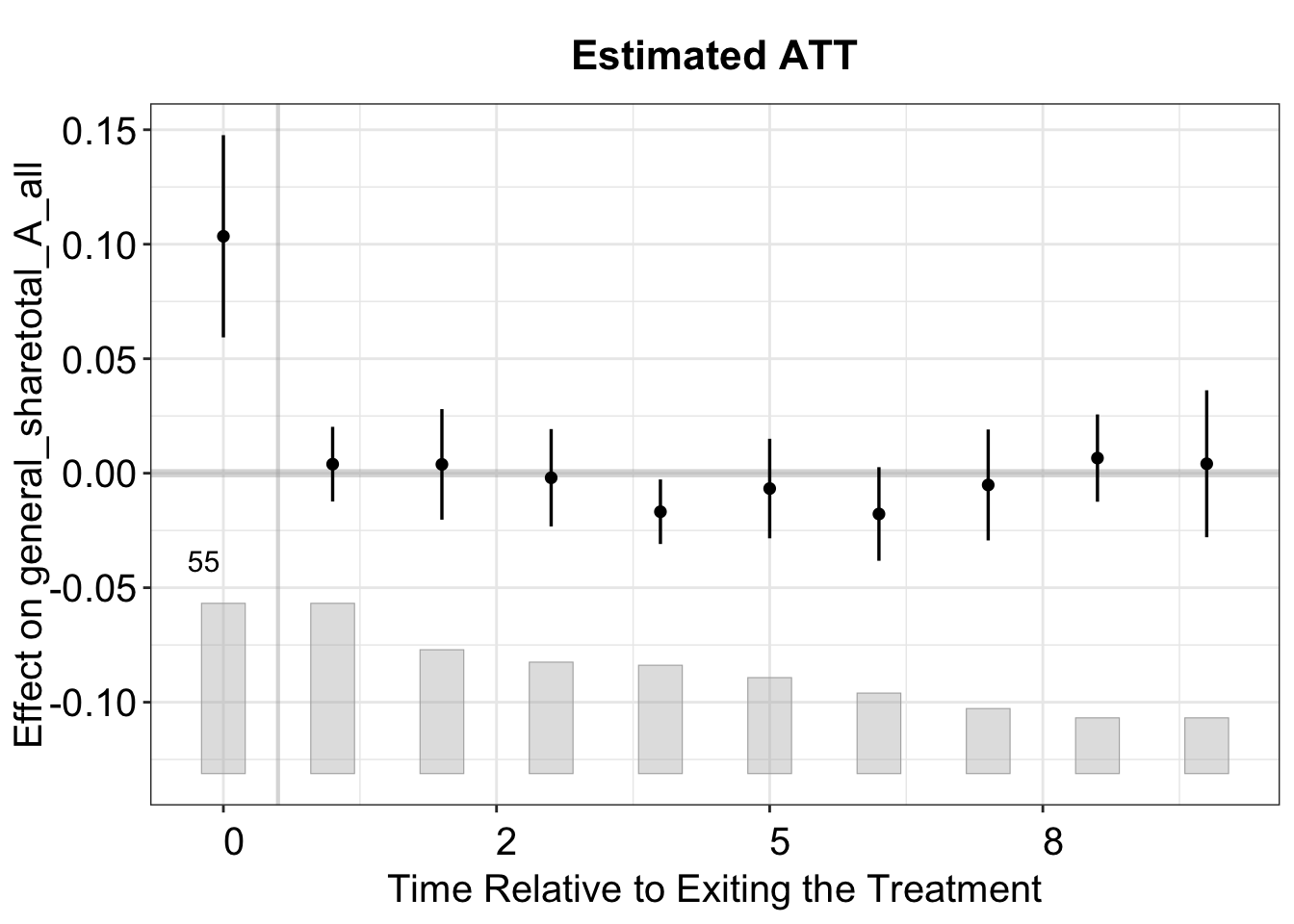
The test for carryover effects examines whether the treatment effect persists after treatment ends. It artificially labels several post-treatment periods as treated and estimates the “placebo effect” in those periods. By setting carryover.period = c(1, 3), we specify a placebo period that includes three post-treatment periods. If the treatment effect is purely contemporaneous (i.e., there are no carryover effects), the test will not reject the null hypothesis. In this application, the average carryover effect is close to zero and statistically indistinguishable from zero.
out_fe_carryover <- fect(Y = y, D = d, X = controls, data = gs2020,
index = c(unit, time), force = "two-way",
parallel = TRUE, se = TRUE, CV = FALSE,
nboots = 1000, carryoverTest = TRUE,
carryover.period = c(1, 3))
#> Some units are totally removed after drop missing values of the outcome or covariates.
#> Parallel computing ...
#> Bootstrapping for uncertainties ...
#> 1000 runs
plot(out_fe_carryover)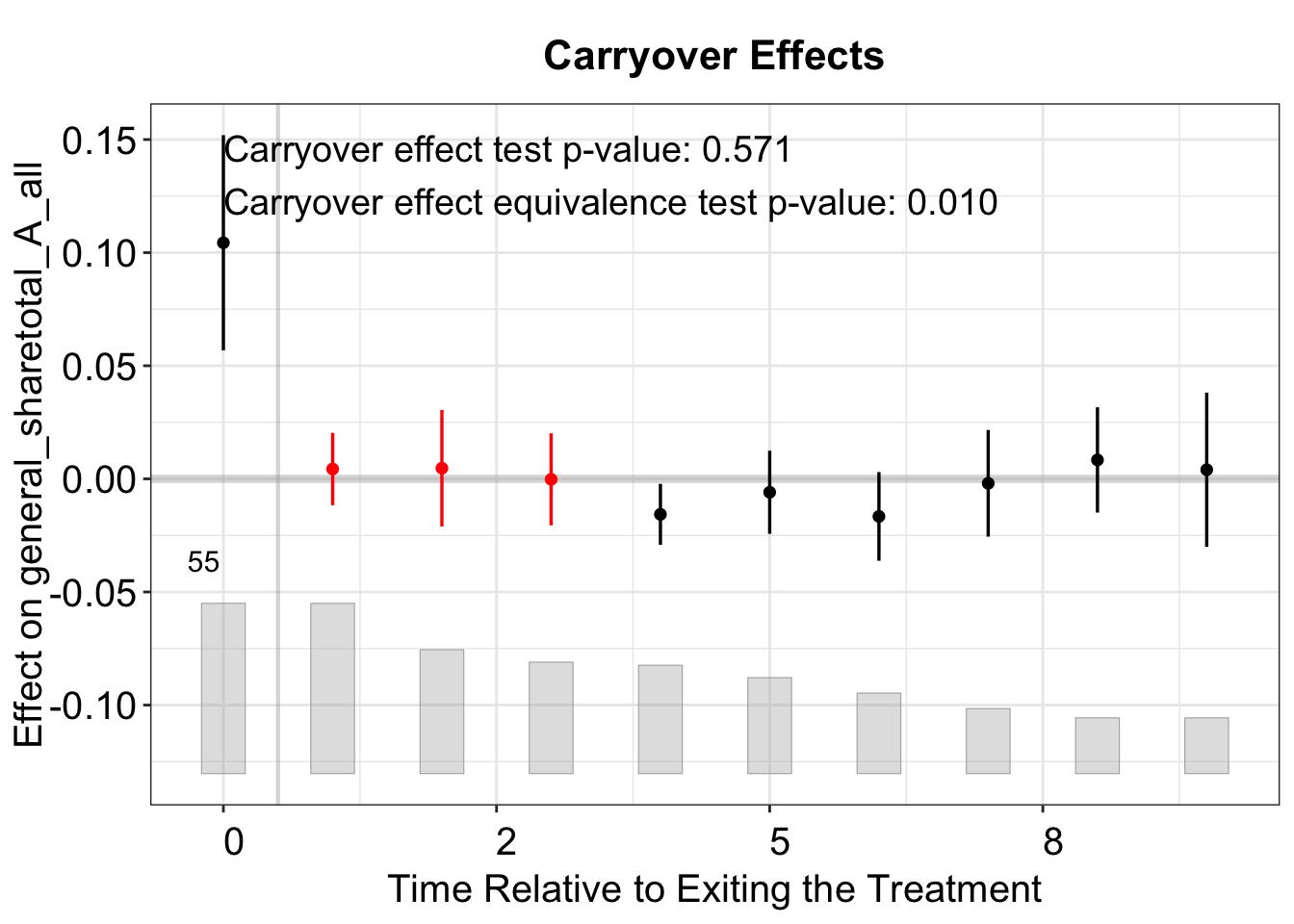
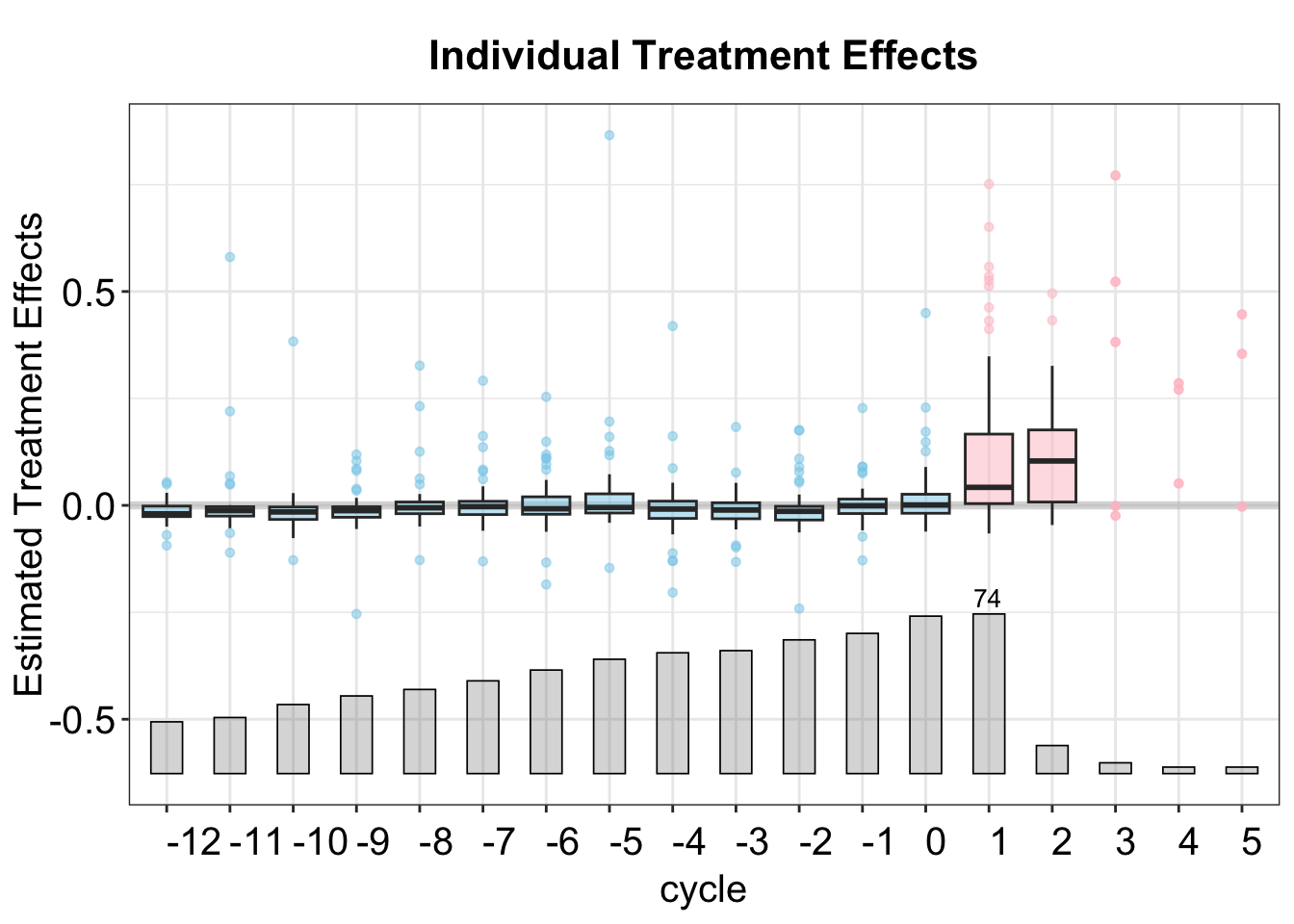
3.5 Effect Heterogenity
We provide two ways to visualize treatment effect heterogeneity: a "box" plot, which shows the distribution of individual treatment effects, and a "calendar" plot, which depicts the ATT conditional on calendar time. We plan to expand this functionality to allow for more pre-treatment covariates soon.
In the box plot, the box in each period represents the range of the middle 50% of the individual effects, while the whiskers show the 2.5%–95% quantiles and the horizontal line represents the median.
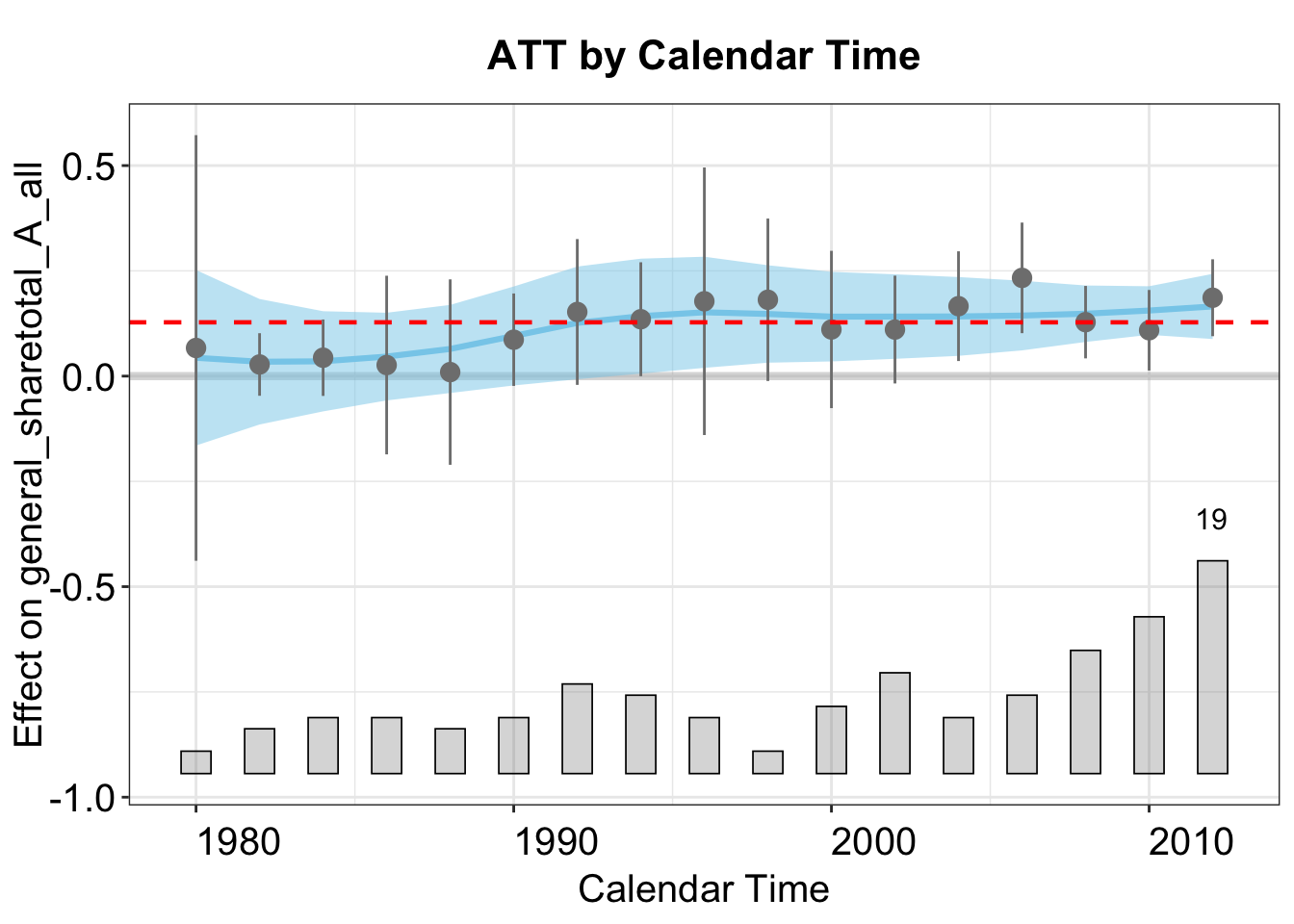
In the calendar plot, the blue ribbon represents a loess fit of the conditional ATT, with 95% confidence intervals.
plot(out, type = "calendar")